Short abstract
The generation of a circadian transcriptional network has led to a better understanding of the gene-expression patterns that regulate the precise 24-hour clock.
Abstract
Circadian rhythms are those biological rhythms that have a periodicity of around 24 hours. Recently, the generation of a circadian transcriptional network - compiled from RNA-expression and promoter-element analysis and phase information - has led to a better understanding of the gene-expression patterns that regulate the precise 24-hour clock.
More than 30 years ago, Seymour Benzer started on the quest for the holy grail of behavioral neuroscience, the elucidation of behavior at a molecular and systems level [1]. In part because of his efforts, this quest is perhaps furthest along in the study of biological rhythms, which in mammals are most clearly manifested in the regulation of a very primitive behavior, the sleep-wake cycle. Elegant genetic and biochemical studies in several species have revealed that the circadian clock that controls such daily rhythms is a cell-autonomous transcriptional/translational feedback loop (reviewed in [2]). In mammals, the master circadian clock resident in the suprachiasmatic nucleus (SCN) of the hypothalamus functions to synchronize other oscillators that drive physiological outputs to a 24-hour rhythm. Despite increasingly refined models of how individual clock components function together as a self-sustaining oscillator, the link between their action, transcriptional oscillations (or clock output), and dependent processes such as physiology and behavior has remained elusive.
Enter systems biology, which can be broadly defined as the integration and synthesis of information from various subfields to inform a biological question [3]. In this field, changes to biological systems are observed at multiple levels under a set of experimental condition(s). Integration of complex data, such as RNA and protein levels together with phenotypes, facilitates the construction of prospective models, which can inform and be informed by experimental data. The methodologies used may include, but are not limited to, transcriptional profiling, differential proteomics, cell-based screening, and whole-organism phenotypic screening [3]. These studies often produce information-rich datasets that necessitate the use of bioinformatics tools to organize and manage the information and to synthesize testable hypotheses.
As a nascent field, many of the initial studies could be viewed as hypothesis-generating experiments. Transcriptional profiling, proteomic screens, and in silico studies in themselves merely capture a snapshot of data coincident with a biological process. As the field has progressed, however, studies have become more refined, and involved the interplay between hypothesis generation and testing.
Systems-level circadian studies
After initial studies in model systems such as Arabidopsis and Drosophila, several authors have applied a popular systems-biology tool - transcriptional profiling - to the study of mammalian circadian transcriptional output [4-8]. Transcriptional profiling, which is usually accomplished using DNA arrays, was employed to identify batteries of genes and biological systems that are controlled by the master clock in the SCN [4,9], as well as rhythms regulated by peripheral clocks in liver, kidney, and heart [4-8]. Although the core circadian activators, Bmal1 and Clock, and the core repressors, the cryptochromes, function analogously in these tissues, their downstream targets vary between different tissues. For example, analysis of rhythmic genes in liver revealed their principal roles to be regulation of metabolism, whereas genes cycling in the SCN were primarily involved in signaling and neurosecretion. These studies also uncovered, to a varying degree, a central battery of genes that are clock-regulated in every tissue and can be thought of as first-order clock-controlled genes (CCGs); some of these, such as Bmal1, the cryptochrome Cry1, the period homolog Per2, and the nuclear receptor Nr1d1 are components of the clock itself [4,6-8]. More recent profiling studies have used genetic models to refine both the roles of these core components and their outputs. Using animals with a functionally deficient locus for several of the known circadian regulators, such as Clock mutant and cryptochrome-deficient Cry1-/- Cry2-/- mice, studies have shown both the specificity and redundancy of various core components and the effect of their deletion on rhythmic behavior and transcription [7].
But this still begs the question of who is regulating whom and, an especially important question for biological timing, when? In a recent report, Ueda et al. [10] have begun to address these very issues by using system-level approaches to explore the network topology of circadian transcriptional output (Figure 1). The authors have collated information from their earlier transcriptional studies and those of others to generate a list of sixteen cycling genes that have also been identified as members of the circadian regulatory machinery. These candidates were then screened in assays in vitro to assess the contribution of their regulatory elements to both cycling activity and to the temporal phase of peak expression. Many of these regulatory elements were themselves targets of other clock genes, reflecting the interlocking nature of circadian feedback loops. Of the sixteen genes explored in detail, nine were found to harbor functional E/E' boxes, targets of the Clock/Bmal1 complex, in their promoter regions, seven had functional DBP/E4BP4-binding elements (D boxes), and six harbored functional RevErbA/ROR-regulatory elements (RREs) [10]. From the promoter information, the transcriptional regulators could be grouped on the basis of the phase of the circadian cycle in which they were transcriptionally most active.
Figure 1.
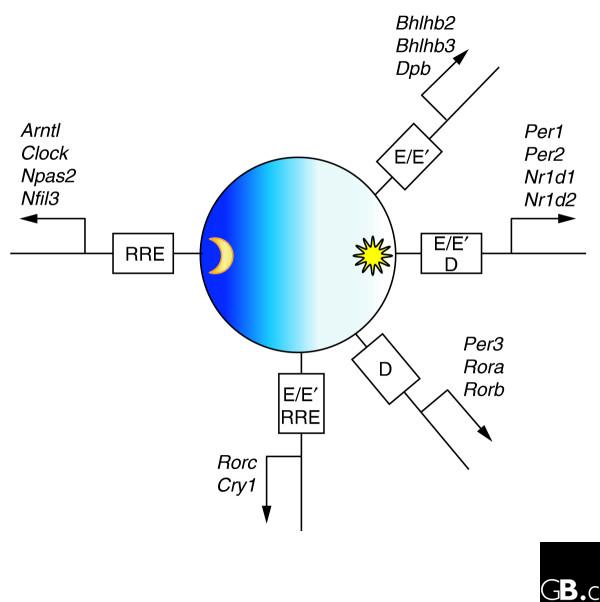
Regulatory elements of mammalian circadian gene expression. A systems-level approach has identified the transcriptional circuit controlling circadian gene expression. The E/E3 box, the DBP/E4BP4-binding element (D box), and the RevErbA/ROR-regulatory element (RRE) are upstream regulatory elements in the genes indicated, and function alone or in combination throughout the 24-hour cycle to generate five phases of gene expression. The genes shown encode the following proteins: Arntl, aryl hydrocarbon receptor nuclear translocator-like protein; Bhlhb2 and Bhlhb3, basic helix-loop-helix domain containing proteins, class B; Clock, circadian locomotor output cycles kaput protein; Cry1, cryptochrome 1; Dpb, D-site albumin promoter-binding protein; Nr1d1 and Nr1d2, nuclear receptor subfamily 1 group D proteins; Npas2, neuronal PAS domain protein 2; Nfil3, nuclear factor, interleukin-3-regulated; Per1-Per3, period homologs; Rora, Rorb and Rorc, retinoic acid receptor-related orphan receptors alpha, beta, and gamma.
An important aspect of circadian biology is the requirement for rhythmic output throughout the entire 24-hour day. How does the clock regulate gene expression throughout the entire day given a limited number of regulatory elements? This is known as the phase-control problem. Ueda et al. [10] elegantly demonstrate how complex phase regulation can be accomplished using combinations of three clock-regulated elements: E/E' boxes, D boxes, and RREs. These experiments show how constructive and destructive interference can be used to generate new phases and amplitudes of circadian transcription. Ueda et al. [10] used in silico methods to construct models that accurately reflect the observations that cycling genes can have low-amplitude or high-amplitude oscillations. The phase of these oscillations can shift forwards or backwards depending on which cohort of genes is regulating their expression. Using this foundation of knowledge, it was possible to construct a model of the circuit of the circadian feedback system. This model was then probed to find the Achilles' heel of the transcriptional circuit underlying the mammalian circadian clock. These experiments supported the proposed model, and concluded that the E/E' box plays the critical role in the regulation of circadian transcription.
The advent of systems biology has allowed the elucidation of biological features such as behavior. Complicated feedback loops can be decoded, allowing the identification of central regulators of the system and those controlling more subtle processes. With respect to circadian behavior, these studies have reinforced the importance of the E/E' box regulators Clock, Bmal1, Cry1 and Cry2. The true importance of these studies, however, lies in the construction of sophisticated models of regulatory systems that can be experimentally tested. The continued application of these approaches, coupled with rigorous experimental testing to confirm prospective modeling, provides a remarkable opportunity to explain how behavior can result from relatively simple transcriptional networks. Benzer's quest continues, and indeed, the magnitude of the task is only now becoming apparent. Despite this, Ueda et al. [10] are leading the way in helping us see how systems biology can shed light on the circadian clock.
References
- Benzer S. Genetic dissection of behavior. Sci Am. 1973;229:24–37. doi: 10.1038/scientificamerican1273-24. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/S0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Ideker T, Galitski T, Hood L. A new approach to decoding life: systems biology. Annu Rev Genomics Hum Genet. 2001;2:343–372. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12:540–550. doi: 10.1016/S0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, Dunlap JC. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol. 2002;12:551–557. doi: 10.1016/S0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- Kita Y, Shiozawa M, Jin W, Majewski RR, Besharse JC, Greene AS, Jacob HJ. Implications of circadian gene expression in kidney, liver and the effects of fasting on pharmacogenomic studies. Pharmacogenetics. 2002;12:55–65. doi: 10.1097/00008571-200201000-00008. [DOI] [PubMed] [Google Scholar]
- Oishi K, Miyazaki K, Kadota K, Kikuno R, Nagase T, Atsumi G, Ohkura N, Azama T, Mesaki M, Yukimasa S, et al. Genome-wide expression analysis of mouse liver reveals CLOCK-regulated circadian output genes. J Biol Chem. 2003;278:41519–41527. doi: 10.1074/jbc.M304564200. [DOI] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]


