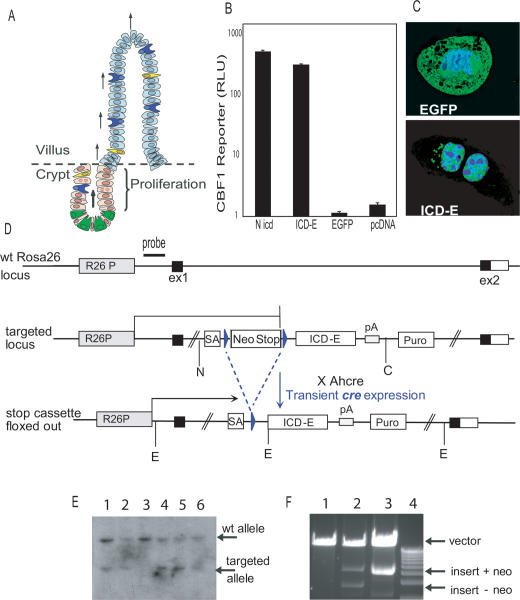
Figure 1.
Construct design, validation, and targeting. (A) Structure of the intestinal epithelium; the cell types shown are enterocytes (pale blue), goblet cells (dark blue), endocrine cells (yellow), Paneth cells (green), and progenitors (pink); arrows indicate the migration of cells from the crypts to the villi. (B) In vitro validation of construct. cDNA encoding mouse Notch 1 cytoplasmic domain (Nicd), ICD-E, EGFP, or empty vector was transfected into CHO cells with a CBF1 DNA-binding motif firefly luciferase reporter vector, and a Renilla luciferase control (Strobl et al. 1997). Error bars indicate SEM. (C) Confocal microscopy showing localization of EGFP and ICD-E (green) in transfected 3T3 cells also stained with DAPI for DNA (blue). (D) Conditional targeting of ICD-E to the Rosa 26 locus. Upstream of the ICD-E fusion a PGK neo pA sequence served as a Stop cassette. This was flanked by LoxP sequences (blue triangles). A splice acceptor (SA) was introduced in front of the floxed Stop cassette to allow normal processing of ICD-E RNA following excision of the Stop cassette by transiently expressed cre from the Ahcre line. The construct was cloned into pRos26-MCS-13 for targeting to the ROSA26 locus. (E) Southern blot analysis of ES cell clones confirmed the targeting of the construct to the ROSA26 locus. The location of the probe used in the Southern blot analysis is shown in D. The upper band corresponds to the EcoRI (E) fragment of the wild-type allele and the lower band to the smaller EcoRI fragment of the targeted allele shown in D. Lanes 1–6 represent ES cell lines CA2, CA3, CB3, CC3, CD1, and CH1, respectively. Mice derived from lines CA2 and CD1 were selected and used in this study; the described phenotype was seen in both lines. (F) In vitro cre-mediated recombination of the targeting construct. The plasmids shown were incubated with or without cre recombinase in vitro, and analyzed after a restriction digest to release the insert from pRosa26-MCS-13. (Lane 1) Linearized pRosa26-MCS-13, no cre incubation. (Lane 2) pRosa26-MCS-13 + insert incubated with cre and subsequently digested with NheI and ClaI (sites N and C, respectively, in D). (Lane 3) pRosa26-MCS-13 + insert digested with NheI and ClaI, no cre incubation. (Lane 4) One-kilobase ladder.