Abstract
Background
Previous studies mostly use single-type features to establish a prediction model. We aim to develop a comprehensive prediction model that effectively identify patients with poor prognosis for single hepatocellular carcinoma (HCC) based on artificial intelligence (AI). Patients and methods: 236 single HCC patients were studied to establish a comprehensive prediction model. We collected the basic information of patients and used AI to extract the features of magnetic resonance (MR) images.
Results
The clinical model based on linear regression (LR) algorithm (AUC: 0.658, 95%CI: 0.5021–0.8137), the radiomics model and deep transfer learning (DTL) model based on light gradient-boosting machine (Light GBM) algorithm (AUC: 0.761, 95%CI: 0.6326–0.8886 and AUC: 0.784, 95%CI: 0.6587–0.9087, respectively) were the optimal prediction models. A comparison revealed that the integrated nomogram had the largest area under the receiver operating characteristic curve (AUC) (all P < 0.05). In the training cohort, the integrated nomogram was predictive of recurrence-free survival (RFS) as well as overall survival (OS) (C-index: 0.735 and 0.712, P < 0.001). In the test cohort, the integrated nomogram also can predict RFS and OS (C-index: 0.718 and 0.740, P < 0.001) in patients.
Conclusion
The integrated nomogram composed of signatures in the prediction models can not only predict the postoperative recurrence of single HCC patients but also stratify the risk of OS after the operation.
Keywords: Artificial intelligence, Hepatocellular carcinoma, Prognosis
1. Introduction
HCC is the most common primary liver cancer [1], and its morbidity and mortality rates are among the highest worldwide [2]. Surgical resection is the first-line treatment currently for patients with early HCC [3]. Advanced HCC or those who are inoperable due to other factors, liver transplantation, radiofrequency ablation, targeted therapy, or immunotherapy is still available [4,5]. However, the survival benefits of the patients are not well improved. Therefore, improving the survival benefits of HCC patients is still one of the challenges in current medicine.
With the completion of the human genome project, the concepts of precision oncology and personalized medicine were born [6]. HCC is a highly genetically heterogeneous tumor [7], which reduces the value of genomic analysis to some extent. Genomic analysis is based on surgical tumor specimens or invasive biopsies. Due to factors such as repeatability and safety, it is difficult to promote and popularize genomic analysis in clinical practice. Radiomics has arisen because of its ability to optimize existing imaging resources. At present, there are no unified definitions of radiomics. It generally means the high-throughput extraction of quantitative features from medical images and further analysis of the extracted features by specific algorithms, to develop prediction models for clinical diagnosis, efficacy evaluation after treatment, prognosis, and other clinical decisions [8]. According to the radiomics hypothesis, the imaging presentation of the tumor contains information reflecting the underlying pathophysiology [9]. Some scholars pointed out that the radiological characteristics are highly correlated with the heterogeneity index [10]. As a non-invasive, relatively safe, and highly repeatable examination method, imaging examination has been widely accepted by patients. For patients with HCC, non-invasive MR imaging examination is of great significance. Because the clinical diagnosis of HCC can be carried out without relying on invasive histopathological examination.
At present, solving complex problems through interdisciplinary cooperation has become the mainstream. The application of AI in medical field is helpful to form research results with practical guiding significance [11]. Deep learning and machine learning commonly used in medical study belong to the AI, a branch of computer science [12]. In the medical field, deep learning has been extensively used recently. Among them, the depth convolutional neural network (DCNN) is an artificial neural network widely used in deep learning, which is usually used for image recognition [13]. The DCNN can automatically extract and learn the deep features of the input data without manual design [14]. AI is likely to improve the comprehensive management of HCC by improving the accuracy of diagnosis, prognosis, and risk prediction of HCC [15]. There are some scholars have successfully established models to distinguish HCC from other space-occupying lesions based on AI [[16], [17], [18]]. Deep learning based on MR images can be used to evaluate the microvascular invasion (MVI) of HCC [[19], [20], [21]], and the grading of HCC before operation [22,23]. Studies have shown that it is effective to develop a prediction model to predict the prognosis of HCC based on MR imaging radiomics [24]. Some scholars have successfully predicted the recurrence of HCC after treatment using deep learning analysis of baseline MR images characteristics [25]. And there are some scholars have established nomograms based on deep learning to predict the postoperative recurrence of HCC [26].
However, previous studies mostly focused on the prediction of the outcome based on single-type features, and few studies established prediction models based on comprehensive clinical features, radiomics features, and DTL features. Based on the above, we attempted to establish a comprehensive prediction model capable of predicting the prognosis of the single HCC using AI. Therefore, we aim to develop a comprehensive predictive model that effectively identifies patients with poor prognosis after curative surgery for the single HCC.
2. Material and methods
2.1. Patient population
Our research has been reviewed and approved by the Hospital Ethics Committee. Patients with a single HCC who visited our hospital from May 2017 to July 2018 were selected. According to the following inclusion and exclusion criteria, a total of 236 patients were included. Inclusion criteria: (1) patient diagnosed with HCC by histopathology; (2) patient undergoing curative hepatectomy; (3) patient with single HCC. Exclusion criteria: (1) patient under 18 years old; (2) patient with transferred liver cancer; (3) patient with other malignant tumors; (4) patient with severe cardiac, pulmonary, hepatic, and renal insufficiency; (5) patient without enhanced MR imaging examination of the liver; (6) patient with preoperative anti-tumor therapy. Our follow-up ended in May 2023. The follow-up methods were the hospital's electronic medical record system and telephone. Recurrence was defined as a recurrence of the tumor diagnosed on imaging.
2.2. MR images acquisition and tumor segmentation
Preoperative MR images of all enrolled patients were downloaded from the hospital's imaging system. The tumor region segmentation process of each MR image was performed twice by a radiologist with more than 10 years of work experience at different times to reduce the segmentation error. First, the unenhanced phase, arterial phase, portal venous phase, and delayed phase of the MR images were selected. And then the maximum tumor area layer was determined in the images of each phase. Finally, the contour edges of the tumors were manually drawn using 3D-Slicer software to obtain the region of interest (ROI).
2.3. Statistical analysis
See the supplementary document 1 for the method of comparing the differences of variables. To identify the independent clinical predictors of 5-year recurrence in HCC patients after the operation, we use the univariate and multivariate logistic regression. The clinical features with a P value < 0.05 were selected as the final features for the construction of the clinical model. We adopt four methods as the means of feature screening, namely Spearman's rank correlation coefficient, intraclass correlation coefficient (ICC), T-test, and the least absolute shrinkage and selection operator (LASSO). Evaluation of prediction performance of prediction model by the accuracy, sensitivity, specificity, negative predictive value (NPV), positive predictive value (PPV), and AUC. Evaluation of calibration efficiency of prediction model by the Calibration curves and Hosmer-Lemeshow test. In addition, the decision curve analysis (DCA) was used to evaluate the clinical practicability of the prediction model. The differences between the AUC were examined by the DeLong test. The survival curve of RFS and OS is identified by Kaplan-Meier analysis, and the differences are compared by the log-rank test. Statistical analysis was performed using Python (version 3.7), MedCalc (version 20.0), and SPSS (version 25.0). P < 0.05 was considered statistically significant.
3. Result
The total number of eligible patients we enrolled was 236. There are 206 males and 30 females. The median follow-up time was 61.75 months (range 8.9–73.9). A total of 158 subjects had a postoperative recurrence until the end of the follow-up. The 1-, 3-, and 5-year OS was 98.7 %, 76.7 %, and 55.1 %, respectively. Two patient cohorts of a training cohort and a test cohort were established by random allocation in a 3:1 allocation ratio (supplementary document 2 and Table S1).
See supplementary document 3 for the calculation method of inflammatory markers and non-invasive scoring systems. Table S2 shows that gender, liver cirrhosis, MVI status, total bilirubin, alkaline phosphatase, albumin-bilirubin (ALBI), and γ-glutamyl transpeptidase-to-lymphocyte ratio (GPR) were associated with 5-year recurrence in the training cohort (P < 0.05). Tumor diameter, alanine aminotransferase (ALT) and aspartate aminotransferase (AST) were associated with 5-year recurrence in the test cohort (P < 0.05).
In the training cohort, the P < 0.05 variables in the univariate logistic regression were gender, liver cirrhosis, MVI status, neutrophil-to-lymphocyte ratio (NLR), and ALBI, which were included in the multivariate logistic regression. Ultimately, we concluded that the three clinical features of gender, liver cirrhosis, and MVI status were risk factors for postoperative recurrence in patients with single HCC (all P < 0.05, Table 1).
Table 1.
Univariate and multivariate logistic analysis in training cohort.
| Variable |
Univariate analysis |
Multivariate analysis |
||
|---|---|---|---|---|
| OR (95%CI) | P | OR (95%CI) | P | |
| Gender (female) | 0.367 (0.153–0.880) | 0.025 | 0.376 (0.143–0.991) | 0.048 |
| Age | 1.014 (0.985–1.044) | 0.337 | ||
| HBsAg (+) | 0.728 (0.334–1.590) | 0.426 | ||
| TNM stage | ||||
| I | 1.000 | |||
| II | 0.911 (0.371–2.239) | 0.839 | ||
| III | 1.082 (0.450–2.603) | 0.860 | ||
| IV | / | 0.999 | ||
| Liver cirrhosis (yes) | 2.026 (1.070–3.834) | 0.030 | 2.148 (1.085–4.251) | 0.028 |
| Cell differentiation | ||||
| poor | 1.000 | |||
| moderate | 1.176 (0.618–2.238) | 0.620 | ||
| well | 1.098 (0.255–4.731) | 0.900 | ||
| MVI status (+) | 2.948 (1.272–6.837) | 0.012 | 3.362 (1.387–8.149) | 0.007 |
| Tumor diameter (cm) | 1.112 (0.999–1.237) | 0.052 | ||
| AFP | 1.000 | 0.619 | ||
| WBC (∗10^9/L) | 1.084 (0.924–1.272) | 0.324 | ||
| Neutrophil (∗10^9/L) | 1.221 (0.978–1.526) | 0.078 | ||
| Lymphocyte (∗10^9/L) | 0.883 (0.543–1.435) | 0.616 | ||
| Monocyte (∗10^9/L) | 3.154 (0.650–15.317) | 0.154 | ||
| Hemoglobin (g/L) | 1.004 (0.985–1.022) | 0.691 | ||
| Platelet (∗10^9/L) | 0.998 (0.994–1.002) | 0.282 | ||
| ALT (U/L) | 1.006 (0.995–1.017) | 0.313 | ||
| AST (U/L) | 1.017 (0.996–1.038) | 0.107 | ||
| Albumin (g/L) | 0.948 (0.880–1.022) | 0.166 | ||
| TBil (μmol/L) | 1.064 (1.000–1.133) | 0.051 | ||
| ALP (U/L) | 1.009 (1.000–1.019) | 0.060 | ||
| γ-GT (U/L) | 1.002 (0.999–1.006) | 0.236 | ||
| PT (S) | 1.178 (0.832–1.669) | 0.356 | ||
| NLR | 1.427 (1.035–1.968) | 0.030 | 1.407 (0.988–2.003) | 0.058 |
| PLR | 1.001 (0.996–1.005) | 0.741 | ||
| PNI | 0.964 (0.913–1.019) | 0.194 | ||
| APRI | 1.448 (0.843–2.486) | 0.180 | ||
| ALBI | 2.640 (1.067–6.530) | 0.036 | 1.882 (0.684–5.178) | 0.221 |
| Fib-4 | 1.145 (0.940–1.395) | 0.178 | ||
| GPR | 1.567 (0.884–2.776) | 0.124 | ||
OR: odd ratio, CI: confidence interval, HBsAg: hepatitis B surface antigen, TNM: tumor node metastasis, MVI: microvascular invasion, AFP: alpha-fetoprotein, WBC: white blood cell, ALT: alanine aminotransferase, AST: aspartate aminotransferase, TBil: total bilirubin, ALP: alkaline phosphatase, γ-GT: γ-glutamyl transpeptidase, PT: prothrombin time, NLR: neutrophil-to-lymphocyte ratio, PLR: platelet-to-lymphocyte ratio, PNI: prognostic nutritional index, APRI: aspartate aminotransferase-to-lymphocyte ratio index, ALBI: albumin-bilirubin, Fib-4: fibrosis 4 score, GPR: γ-glutamyl transpeptidase-to-lymphocyte ratio.
Different acquisition protocols may cause differences in the distances between different voxels in the MR images. The pixels of the rendered ROIs were adjusted using a fixed resolution (1mm × 1mm × 1 mm) before the selection of radiomics features to reduce the impact of differences on feature extraction. The geometry features, intensity features, and texture features extracted from each ROI are collectively referred to as handcrafted features (supplementary document 4). The entire process is based on PyRadiomics (supplementary document 5), which meets the ISBI standard. We used several different radiomics algorithms (supplementary document 6). Finally, we extracted 1834 features including 14 geometry features, 360 intensity features, and 1460 texture features from each ROI.
In order to extract the features of DTL, the gray values for each ROI were first normalized to a minimum-to-maximum conversion in the range of [−1, 1]. Using nearest neighbor interpolation, the cropped sub-region image is then adjusted to 224 × 224 pixels. In addition, the cosine annealing algorithm is used to improve the generalization of the model (supplementary document 7). We select the fine-tuned ResNet50 model as the DCNN model to extract the DTL feature, which has been pre-trained on the ImageNet ILSVRC-2012 dataset (supplementary document 8). We know that the latter layer of the general network has stronger semantic expression ability. So, the penultimate layer, namely the avgpooling layer, is selected to extract the feature, which extracts 2048-dimensional features. To understand the features related to model classification in ROIs, we use the gradient-weighted class activation mapping, which is also convenient for studying the interpretability of the DTL model. It can be seen that these thermal diagrams highlight those local areas in the particular category identified by the model (Fig. S1).
To improve the stability of the features used for recurrence prediction, avoid over-fitting of the features, and improve the generalization of the prediction model, we have screened the extracted radiomics features and DTL features (supplementary document 9). After feature selection, different machine learning algorithms are used to establish prediction models for clinical features, the selected radiomics features, and DTL features. We developed predictive models by several different machine learning algorithms: LR, support vector machine (SVM), Extra-Trees, eXtreme Gradient Boosting (XG Boost), and Light GBM. To obtain a prediction model with optimal performance, we use the Grid-Search algorithm to search for optimal hyperparameters for each machine learning algorithm.
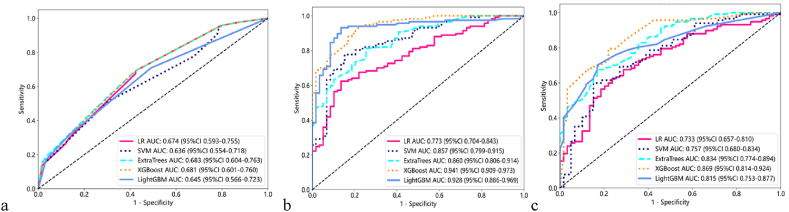
Next, we use different machine learning algorithms to construct clinical models in the training cohort, respectively (Fig. 1a). Similarly, for the radiomics features and DTL features selected in the training cohort, different machine learning algorithms are used to respectively construct the radiomics models (Fig. 1b) and DTL models (Fig. 1c). Detailed results for the performance of different predictive models using different machine learning algorithms are presented in Table S3. We uniformly selected the prediction model with the largest AUC in the test cohort as the optimal prediction model for the postoperative recurrence of patients with HCC. The clinical model based on the LR algorithm, the radiomics model, and the DTL model based on the Light GBM algorithm are the optimal prediction models (Table 2). In the test cohort, the ROC curves of all prediction models are shown in Fig. S2.
Fig. 1.
The ROC curves of clinical models(a), radiomics models(b) and DTL models(c) in the training cohort.
ROC: receiver operating characteristic, DTL: deep transfer learning, LR: linear regression, SVM: support vector machine, XG Boost: eXtreme Gradient Boosting, Light GBM: light gradient-boosting machine.
Table 2.
Evaluation of performance on Clinical model, radiomics model and DTL model in test cohort.
| Algorithm | AUC | 95 % CI | Accuracy | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|---|
| Clinical model | |||||||
| LR | 0.658 | 0.5021–0.8137 | 0.650 | 0.659 | 0.750 | 0.794 | 0.462 |
| SVM | 0.474 | 0.3169–0.6318 | 0.717 | 0.976 | 0.188 | 0.714 | 0.750 |
| Extra-Trees | 0.612 | 0.4445–0.7788 | 0.650 | 0.659 | 0.750 | 0.794 | 0.462 |
| XGBoost | 0.658 | 0.5021–0.8137 | 0.650 | 0.659 | 0.750 | 0.794 | 0.462 |
| Light GBM | 0.626 | 0.4773–0.7756 | 0.633 | 0.659 | 0.688 | 0.771 | 0.440 |
| Radiomics model | |||||||
| LR | 0.721 | 0.5872–0.8557 | 0.700 | 0.683 | 0.737 | 0.848 | 0.519 |
| SVM | 0.685 | 0.5404–0.8306 | 0.650 | 0.561 | 0.842 | 0.885 | 0.471 |
| Extra-Trees | 0.729 | 0.5980–0.8603 | 0.667 | 0.585 | 0.842 | 0.889 | 0.485 |
| XGBoost | 0.653 | 0.5044–0.8024 | 0.667 | 0.634 | 0.778 | 0.839 | 0.483 |
| Light GBM | 0.761 | 0.6326–0.8886 | 0.700 | 0.610 | 0.895 | 0.926 | 0.515 |
| DTL model | |||||||
| LR | 0.714 | 0.5631–0.8644 | 0.783 | 0.878 | 0.579 | 0.818 | 0.687 |
| SVM | 0.701 | 0.5523–0.8495 | 0.717 | 0.780 | 0.579 | 0.800 | 0.550 |
| Extra-Trees | 0.724 | 0.5801–0.8679 | 0.750 | 0.829 | 0.579 | 0.810 | 0.611 |
| XGBoost | 0.767 | 0.6299–0.9041 | 0.783 | 0.878 | 0.611 | 0.818 | 0.687 |
| Light GBM | 0.784 | 0.6587–0.9087 | 0.783 | 0.829 | 0.684 | 0.850 | 0.650 |
AUC: area under the receiver operating characteristic curve, CI: confidence interval, PPV: positive predictive value, NPV: negative predictive value, DTL: deep transfer learning, LR: linear regression, SVM: support vector machine, XG Boost: eXtreme Gradient Boosting, Light GBM: light gradient-boosting machine.
To improve the prediction efficiency, we further attempted to establish a nomogram that integrates the above three models for predicting the recurrence of HCC after surgery (Fig. 2a), and compared the individual prediction capabilities of the four predictive models (Table S4). A comparison revealed that the integrated nomogram had the largest AUC (all P < 0.05; Table 3). The calibration curves of the different model analyses showed the integrated nomogram had a relatively good fit in the training cohort (Fig. 2b). The decision curves analysis both indicated that the integrated nomogram could add more benefit to patients than the clinical model, radiomics model, and DTL model (Fig. 2c). We also performed the DeLong test (Table S5), the calibration curves, and the decision curves (Fig. S3) in the test cohort. Although the results were not as ideal as those in the training cohort, they were still of scientific value.
Fig. 2.
Integrated nomogram (a), Calibration curves (b), and Decision curve analysis (c) for guiding recurrence in training cohort.
DTL: deep transfer learning.
Table 3.
Delong test of different models in training cohort.
| Prediction models | P |
|---|---|
| Clinical model Vs integrated Nomogram | <0.05 |
| Radiomics model Vs integrated Nomogram | <0.05 |
| DTL model Vs integrated Nomogram | <0.05 |
DTL: deep transfer learning.
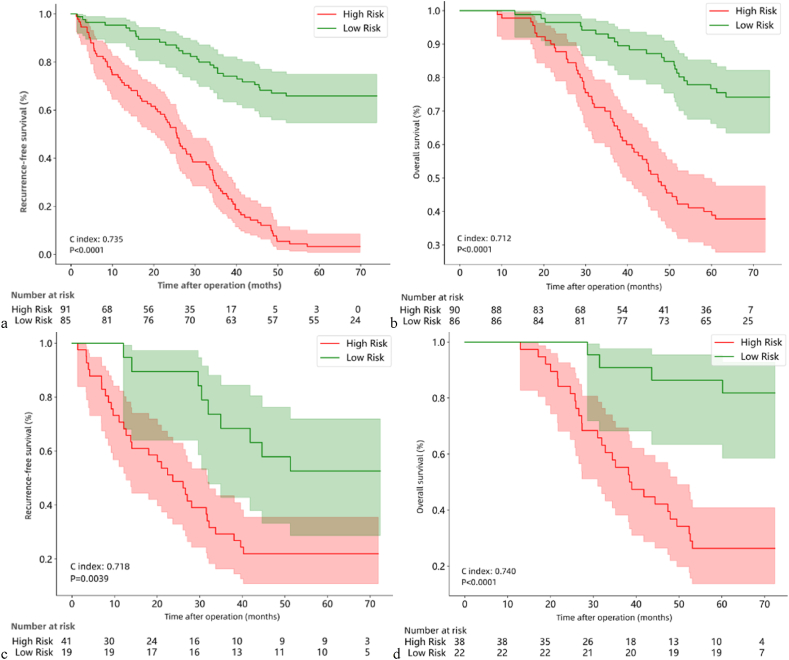
We evaluated the predictive value of the integrated nomogram for RFS and OS in patients using Cox proportional hazards model. High risk was defined for patients with HR > 1 and low risk for patients with HR < 1. The c-index is used to evaluate the prediction performance (Table S6). In the training cohort, the integrated nomogram was predictive of RFS (Fig. 3a, C-index 0.735, P < 0.0001) as well as OS (Fig. 3b, C-index 0.712, P < 0.0001). Next, we validate the integrated nomogram is still able to predict RFS (Fig. 3c, C-index 0.718, P = 0.0039) and OS (Fig. 3d, C-index 0.740, P < 0.0001) in patients in the test cohort.
Fig. 3.
Kaplan-Meier curves of RFS (a) and OS (b) in the training cohort, and Kaplan-Meier curves of RFS (c) and OS (d) in the test cohort.
RFS: recurrence free survival; OS: overall survival.
4. Discussion
Many studies have shown that compared with the traditional prediction model, the prediction model constructed by AI has higher accuracy [27,28]. Comparing to the radiomics model, some studies have pointed out that the deep learning model has better performance [14]. Therefore, the prediction performances of different models are not compared in our study. Based on the results of previous studies, nomograms perform well in the evaluation of prognosis for a variety of cancers, including HCC [29,30,31,32]. So, we combined the clinical signatures, radiomics signatures, and DTL signatures to establish a visualized integrated nomogram. Our results show that the integrated nomogram can not only predict the recurrence of single HCC after the operation but also predict the RFS and OS of single HCC. The innovation of our research consists in the use of multidisciplinary cross-integration to synthesize three kinds of features to improve the accuracy of prediction.
First, radiomics features and DTL features are extracted from the preoperative multi-phase MR images of HCC patients. The crafted signatures in the radiomics model are extracted using machine learning algorithm, while the DTL signatures in the DTL model are extracted using DCNN. After the crafted signatures and DTL signatures extraction is completed, the extracted signatures are selected using Spearman's rank correlation coefficient, ICC, T-test, and the LASSO methods. After the selection, LR, SVM, Extra-Trees, XG Boost, and Light GBM are used to establish the prediction model of the selected clinical signatures, crafted signatures, and DTL signatures, respectively. Then, the signatures of the established prediction model are visualized as an integrated nomogram. The results showed that an integrated nomogram could also predict the recurrence of HCC five years after the operation, and its performance was superior to any single prediction model. Finally, we used the integrated nomogram to predict the RFS and OS of HCC after surgery.
There are several studies have pointed out that NLR, platelet-to-lymphocyte ratio (PLR), and GPR are closely related to the prognosis of HCC [[33], [34], [35]]. Aspartate aminotransferase-to-lymphocyte ratio index (APRI) and fibrosis 4 score (Fib-4) have certain value in the prediction of liver cirrhosis-related adverse events, which has also been confirmed by many scholars [36,37], Some studies have also shown that they have clinical value in the prognosis of HCC [38]. Therefore, when we established the clinical model for predicting the recurrence of HCC five years after the operation, in addition to considering the common clinical characteristics such as gender, age, tumor node metastasis stage, cell differentiation, and MVI status, and basic test items such as routine blood test, liver function, and alpha-fetoprotein, we also considered inflammatory markers such as NLR, PLR, and GPR, as well as non-invasive scoring systems such as APRI and Fib-4.
In our study, only gender, liver cirrhosis, and MVI status were independent risk factors for the recurrence of HCC five years after surgery. The incidence of HBV-associated HCC is significantly higher in men than in women [39]. Studies have pointed out that there are also gender differences, and the prognosis of female patients is better than that of male patients in the prognosis of HCC [40]. Our results suggest that there are also gender differences in the risk of recurrence five years after the operation. It is generally believed that liver cirrhosis is a risk factor for HCC [41]. Here, our findings suggest that liver cirrhosis is also an independent risk factor for the recurrence of HCC five years after the operation. MVI positive is currently recognized as an important indicator that increases the risk of local invasion and distant metastasis of HCC [42]. Our results are also consistent with this view.
As early as 2013, Choi et al. discovered the value of imaging markers by studying the gadoxetic acid-enhanced MR Images of HCC, which have potential predictive value for postoperative tumor recurrence [43]. In recent years, more and more scholars have conducted a series of medical research based on preoperative MR images of HCC. The radiomics model established by Kim et al. based on preoperative MR images can predict the recurrence in HCC patients after operation [44]. Chong et al. established a nomogram to predict the MVI status and prognosis of a single HCC with a diameter <5 cm before operation based on a radiomics model [45]. Kucukkaya et al. showed that it is feasible to predict the recurrence of early HCC from MR images before treatment by using a deep learning algorithm [25]. Zhang et al. prospectively developed a nomogram based on preoperative MR images to predict the recurrence of HCC one year after the operation [46]. Although the methods used are different, all the above studies show that preoperative MR images can effectively predict the prognosis of HCC.
Our research has reached a similar conclusion. Besides, our results also show that in addition to predicting postoperative tumor recurrence, preoperative MR images can also predict postoperative survival of HCC. Moreover, unlike most previous studies, we use multi-parameter MR images. Extracting imaging features from different MR image sequences can provide more abundant tumor biological information, so as to evaluate tumor features more accurately [47].
Due to the lack of a gold standard for evaluating radiomics, we will not compare and evaluate the performance of our prediction model with that of other research models here. We choose a 2D tumor region to represent the whole tumor. Although its representativeness may not be as that of 3D tumor region in general cognition, 2D tumor region segmentation has the advantages of simple operation and small overall error. Moreover, the process of 3D tumor region analysis is more complicated and takes longer.
The advantages of AI in the medical field are obvious, and in the future, it is likely to become an important tool for medical development [48]. But we can't ignore the existing unresolved limitations. Here, we briefly discuss the limitations of establishing a prediction model based on AI. First of all, the model established by AI depends on the size and diversity of samples, which has certain influence on the promotion of the model. Secondly, it is still difficult to fully explain the prediction model based on AI, which leads to the decline of the credibility of the model. Third, there is a lack of standardization of analytical tools. Fourthly, there is a lack of means to automatically segment the target area, because manual segmentation will inevitably lead to human errors. If the limitations can be effectively solved, the individualized treatment and management of HCC may be further improved with the help of MR imaging radiomics based on AI.
Our research also has the following limitations: 1. Lack of external verification queues; 2. Single-center small sample research may lead to limited universality of data; 3. Retrospective studies may cause bias in data selection, which cannot fully reflect the population of the research object. In the future, relevant prospective studies are needed to further verify our prediction model.
5. Conclusion
We established the prediction models that can be used in the clinic to predict the recurrence of single HCC five years after the operation. The integrated nomogram composed of signatures in the prediction models can not only predict the postoperative recurrence of single HCC patients but also stratify the risk of OS after the operation. Our research results provide a reference for individualized treatment and management of HCC.
CRediT authorship contribution statement
Jing Zhou: Writing – review & editing, Writing – original draft, Visualization, Validation, Software, Methodology, Investigation, Formal analysis, Conceptualization. Daofeng Yang: Supervision, Project administration, Data curation. Hao Tang: Writing – review & editing, Visualization, Software, Resources.
Informed consent statement
Patient consent was waived due to this is an observational and retrospective study, as approved by the Institutional Review Board.
Data availability statement
The data underlying this article are aviliable on reasonable request from the first author.
Institutional Review Board statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology (protocol code: TJ-IRB202402017; data of approval: 2 Feb 2024).
Funding
The authors declare no funding.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
No.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2025.e41735.
Contributor Information
Daofeng Yang, Email: yangdaofeng@aliyun.com.
Hao Tang, Email: haotangtjh@163.com.
Appendix A. Supplementary data
The following a the supplementary data to this article:
References
- 1.Zhou J., et al. Guidelines for the diagnosis and treatment of hepatocellular carcinoma (2019 edition) Liver Cancer. 2020;9(6):682–720. doi: 10.1159/000509424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sung H., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca - Cancer J. Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Omata M., et al. Asia-Pacific clinical practice guidelines on the management of hepatocellular carcinoma: a 2017 update. Hepatol Int. 2017;11(4):317–370. doi: 10.1007/s12072-017-9799-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torimura T., Iwamoto H. Treatment and the prognosis of hepatocellular carcinoma in Asia. Liver Int. 2022;42(9):2042–2054. doi: 10.1111/liv.15130. [DOI] [PubMed] [Google Scholar]
- 5.Vogel A., et al. Hepatocellular carcinoma. Lancet. 2022;400(10360):1345–1362. doi: 10.1016/s0140-6736(22)01200-4. [DOI] [PubMed] [Google Scholar]
- 6.Prasad V., Fojo T., Brada M. Precision oncology: origins, optimism, and potential. Lancet Oncol. 2016;17(2):e81–e86. doi: 10.1016/s1470-2045(15)00620-8. [DOI] [PubMed] [Google Scholar]
- 7.Rebouissou S., Nault J.C. Advances in molecular classification and precision oncology in hepatocellular carcinoma. J. Hepatol. 2020;72(2):215–229. doi: 10.1016/j.jhep.2019.08.017. [DOI] [PubMed] [Google Scholar]
- 8.Mayerhoefer M.E., et al. Introduction to radiomics. J. Nucl. Med. 2020;61(4):488–495. doi: 10.2967/jnumed.118.222893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gillies R.J., Kinahan P.E., Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278(2):563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moon S.H., et al. Correlations between metabolic texture features, genetic heterogeneity, and mutation burden in patients with lung cancer. Eur J Nucl Med Mol Imaging. 2019;46(2):446–454. doi: 10.1007/s00259-018-4138-5. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y.P., et al. Artificial intelligence-driven radiomics study in cancer: the role of feature engineering and modeling. Mil Med Res. 2023;10(1):22. doi: 10.1186/s40779-023-00458-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tran K.A., et al. Deep learning in cancer diagnosis, prognosis and treatment selection. Genome Med. 2021;13(1):152. doi: 10.1186/s13073-021-00968-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Litjens G., et al. A survey on deep learning in medical image analysis. Med. Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Li Y., et al. Molecular subtyping of diffuse gliomas using magnetic resonance imaging: comparison and correlation between radiomics and deep learning. Eur. Radiol. 2022;32(2):747–758. doi: 10.1007/s00330-021-08237-6. [DOI] [PubMed] [Google Scholar]
- 15.Calderaro J., et al. Artificial intelligence for the prevention and clinical management of hepatocellular carcinoma. J. Hepatol. 2022;76(6):1348–1361. doi: 10.1016/j.jhep.2022.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jansen M.J.A., et al. Automatic classification of focal liver lesions based on MRI and risk factors. PLoS One. 2019;14(5) doi: 10.1371/journal.pone.0217053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamm C.A., et al. Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi-phasic MRI. Eur. Radiol. 2019;29(7):3338–3347. doi: 10.1007/s00330-019-06205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhen S.H., et al. Deep learning for accurate diagnosis of liver tumor based on magnetic resonance imaging and clinical data. Front. Oncol. 2020;10:680. doi: 10.3389/fonc.2020.00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y., et al. Deep learning with 3D convolutional neural network for noninvasive prediction of microvascular invasion in hepatocellular carcinoma. J Magn Reson Imaging. 2021;54(1):134–143. doi: 10.1002/jmri.27538. [DOI] [PubMed] [Google Scholar]
- 20.Wang G., et al. Prediction of microvascular invasion of hepatocellular carcinoma based on preoperative diffusion-weighted MR using deep learning. Acad. Radiol. 2021;28(Suppl 1):S118–s127. doi: 10.1016/j.acra.2020.11.014. [DOI] [PubMed] [Google Scholar]
- 21.Zhou W., et al. Prediction of microvascular invasion of hepatocellular carcinoma based on contrast-enhanced MR and 3D convolutional neural networks. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.588010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Q., et al. Grading of hepatocellular carcinoma using 3D SE-DenseNet in dynamic enhanced MR images. Comput. Biol. Med. 2019;107:47–57. doi: 10.1016/j.compbiomed.2019.01.026. [DOI] [PubMed] [Google Scholar]
- 23.Li S., et al. Attention guided discriminative feature learning and adaptive fusion for grading hepatocellular carcinoma with Contrast-enhanced MR. Comput Med Imaging Graph. 2022;97 doi: 10.1016/j.compmedimag.2022.102050. [DOI] [PubMed] [Google Scholar]
- 24.Harding-Theobald E., et al. Systematic review: radiomics for the diagnosis and prognosis of hepatocellular carcinoma. Aliment. Pharmacol. Ther. 2021;54(7):890–901. doi: 10.1111/apt.16563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kucukkaya A.S., et al. Predicting tumor recurrence on baseline MR imaging in patients with early-stage hepatocellular carcinoma using deep machine learning. Sci. Rep. 2023;13(1):7579. doi: 10.1038/s41598-023-34439-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan M., et al. Deep learning nomogram based on Gd-EOB-DTPA MRI for predicting early recurrence in hepatocellular carcinoma after hepatectomy. Eur. Radiol. 2023 doi: 10.1007/s00330-023-09419-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singal A.G., et al. Machine learning algorithms outperform conventional regression models in predicting development of hepatocellular carcinoma. Am. J. Gastroenterol. 2013;108(11):1723–1730. doi: 10.1038/ajg.2013.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ioannou G.N., et al. Assessment of a deep learning model to predict hepatocellular carcinoma in patients with hepatitis C cirrhosis. JAMA Netw. Open. 2020;3(9) doi: 10.1001/jamanetworkopen.2020.15626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong L., et al. A deep learning-based radiomic nomogram for prognosis and treatment decision in advanced nasopharyngeal carcinoma: a multicentre study. EBioMedicine. 2021;70 doi: 10.1016/j.ebiom.2021.103522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Y.Q., et al. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J. Clin. Oncol. 2016;34(18):2157–2164. doi: 10.1200/jco.2015.65.9128. [DOI] [PubMed] [Google Scholar]
- 31.Tang L.Q., et al. Establishment and validation of prognostic nomograms for endemic nasopharyngeal carcinoma. J Natl Cancer Inst. 2016;108(1) doi: 10.1093/jnci/djv291. [DOI] [PubMed] [Google Scholar]
- 32.Tang M., et al. Nomogram development and validation to predict hepatocellular carcinoma tumor behavior by preoperative gadoxetic acid-enhanced MRI. Eur. Radiol. 2021;31(11):8615–8627. doi: 10.1007/s00330-021-07941-7. [DOI] [PubMed] [Google Scholar]
- 33.Schobert I.T., et al. Neutrophil-to-lymphocyte and platelet-to-lymphocyte ratios as predictors of tumor response in hepatocellular carcinoma after DEB-TACE. Eur. Radiol. 2020;30(10):5663–5673. doi: 10.1007/s00330-020-06931-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J., et al. A new model based inflammatory index and tumor burden score (TBS) to predict the recurrence of hepatocellular carcinoma (HCC) after liver resection. Sci. Rep. 2022;12(1):8670. doi: 10.1038/s41598-022-12518-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J., et al. Development and validation of a prognostic model based on the albumin-to-fibrinogen ratio (AFR) and gamma-glutamyl transpeptidase-to-platelet ratio (GPR) in hepatocellular carcinoma patients. Clin. Chim. Acta. 2020;511:107–116. doi: 10.1016/j.cca.2020.09.038. [DOI] [PubMed] [Google Scholar]
- 36.Younes R., et al. Long-term outcomes and predictive ability of non-invasive scoring systems in patients with non-alcoholic fatty liver disease. J. Hepatol. 2021;75(4):786–794. doi: 10.1016/j.jhep.2021.05.008. [DOI] [PubMed] [Google Scholar]
- 37.Xiao G., et al. Diagnostic accuracy of APRI and FIB-4 for predicting hepatitis B virus-related liver fibrosis accompanied with hepatocellular carcinoma. Dig. Liver Dis. 2016;48(10):1220–1226. doi: 10.1016/j.dld.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 38.Sonohara F., et al. Comparison of non-invasive liver reserve and fibrosis models: implications for surgery and prognosis for hepatocellular carcinoma. Hepatol. Res. 2019;49(11):1305–1315. doi: 10.1111/hepr.13400. [DOI] [PubMed] [Google Scholar]
- 39.Zheng B., et al. Gender disparity in hepatocellular carcinoma (HCC): multiple underlying mechanisms. Sci. China Life Sci. 2017;60(6):575–584. doi: 10.1007/s11427-016-9043-9. [DOI] [PubMed] [Google Scholar]
- 40.Rich N.E., et al. Sex disparities in presentation and prognosis of 1110 patients with hepatocellular carcinoma. Aliment. Pharmacol. Ther. 2020;52(4):701–709. doi: 10.1111/apt.15917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marrero J.A. Surveillance for hepatocellular carcinoma. Clin. Liver Dis. 2020;24(4):611–621. doi: 10.1016/j.cld.2020.07.013. [DOI] [PubMed] [Google Scholar]
- 42.Zhang X., et al. Significance of presence of microvascular invasion in specimens obtained after surgical treatment of hepatocellular carcinoma. J. Gastroenterol. Hepatol. 2018;33(2):347–354. doi: 10.1111/jgh.13843. [DOI] [PubMed] [Google Scholar]
- 43.Choi J.W., et al. Hepatocellular carcinoma: imaging patterns on gadoxetic acid-enhanced MR Images and their value as an imaging biomarker. Radiology. 2013;267(3):776–786. doi: 10.1148/radiol.13120775. [DOI] [PubMed] [Google Scholar]
- 44.Kim S., et al. Radiomics on gadoxetic acid-enhanced magnetic resonance imaging for prediction of postoperative early and late recurrence of single hepatocellular carcinoma. Clin. Cancer Res. 2019;25(13):3847–3855. doi: 10.1158/1078-0432.Ccr-18-2861. [DOI] [PubMed] [Google Scholar]
- 45.Chong H.H., et al. Multi-scale and multi-parametric radiomics of gadoxetate disodium-enhanced MRI predicts microvascular invasion and outcome in patients with solitary hepatocellular carcinoma ≤ 5 cm. Eur. Radiol. 2021;31(7):4824–4838. doi: 10.1007/s00330-020-07601-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Z., et al. Hepatocellular carcinoma: radiomics nomogram on gadoxetic acid-enhanced MR imaging for early postoperative recurrence prediction. Cancer Imag. 2019;19(1):22. doi: 10.1186/s40644-019-0209-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Z., et al. Self-supervised tumor segmentation and prognosis prediction in osteosarcoma using multiparametric MRI and clinical characteristics. Comput Methods Programs Biomed. 2024;244 doi: 10.1016/j.cmpb.2023.107974. [DOI] [PubMed] [Google Scholar]
- 48.Alabi R.O., et al. Artificial intelligence-driven radiomics in head and neck cancer: current status and future prospects. Int J Med Inform. 2024;188 doi: 10.1016/j.ijmedinf.2024.105464. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are aviliable on reasonable request from the first author.