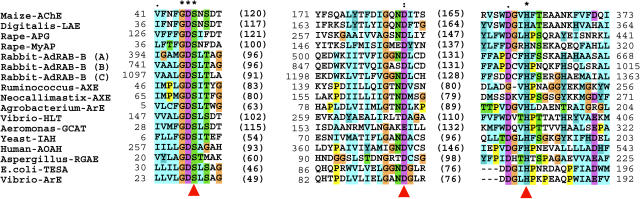
Figure 6.
Alignment of the amino acid sequences of maize AChE and selected members of the GDSL lipase family. The alignment was generated by a ClustalX analysis. Dashes indicate gaps introduced to improve the alignment. Numbers in brackets indicate intervening amino acid numbers, and the numbers without brackets indicate the positions in the sequences. The putative catalytic triad Ser/Asp (Glu)/His residues in the GDSL family enzymes, which were presumed previously (reference in Upton and Buckley, 1995), are indicated by arrowheads. The abbreviations and accession numbers of the cDNA nucleotide sequences used for this analysis are follows: Maize-AChE, Zea mays AChE (determined in this study); Digitalis-LAE, Digitalis lanata lanatoside 15′-O-acetylesterase (AJ011567); Rape-APG, Brassica napus anter-specific Pro-rich protein APG (P40603); Rape-MyAP, Brassica napus myrosinase-associated protein (U39289); Rabbit-AdRAB-B, Oryctolagus cuniculus phospholipase (Z12841); Ruminococcus-AXE, Ruminococcus flavefaciens acetyl xylan esterase (AJ238716); Neocallimastix-AXE, Neocallimastix patriciarum acetylxylan esterase bnaIII (U66253); Agrobacterium-ArE, Agrobacterium radiobacter arylesterase (AF044683); Vibrio-HLT, Vibrio parahaemolyticus thermolabile hemolysin (M36437); Aeromonas-GCAT, Aeromonas hydrophila glycerophospholipid-cholesterol acyltransferase (X07279); Yeast-IAH, Saccharomyces cerevisiae isoamyl acetate hydrolytic enzyme (X92662); Human-AOAH, Homo sapiens acyloxyacyl hydrolase (BC025698); Aspergillus-RGAE, Aspergillus aculeatus rhamnogalacturon acetylesterase (X89714); E. coli-TESA, Escherichia coli acyl-coA thioesterase I (L06182); Vibrio-ArE, Vibrio mimicus arylesterase (X71116).