Figure 3.
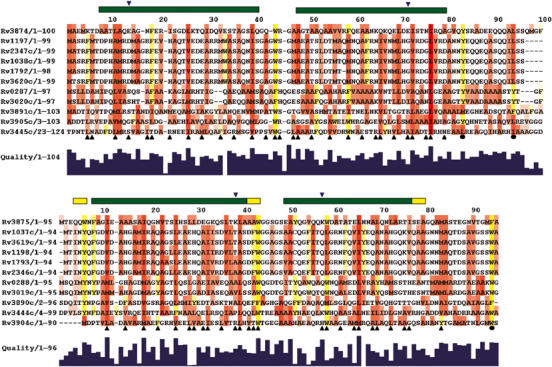
Conservation of the amino-acid sequences of the CFP-10- (top) and ESAT-6- (bottom) related proteins from M. tuberculosis. Aliphatic residues with hydrophobic side chains (Leu, Ile, Val, Met and Ala) are highlighted in red and aromatic residues (Phe, Tyr and Trp) in yellow. The extent of sequence conservation is indicated by the histogram quality score and for highlighted residues the greater the conservation, the more intense the colour. Residues forming the interface between CFP-10 and ESAT-6 are indicated by upright black triangular symbols and conserved hydrophobic residues in the C-terminal arms of the proteins by closed circles. Similarly, the locations of four residues forming two intermolecular salt bridges are indicated by down-turned blue triangles. The positions of helices in the structure of the complex are shown by bars above the sequences (α in green and 310 in yellow). The sequences were aligned using ClustalW, with a standard Blosum30 scoring matrix, a gap opening penalty of 10 and a gap extension penalty of 1 (Thompson et al, 1997).
