Abstract
In multiple systems, impaired proteolysis associated with the loss of the hemostatic factor plasminogen (Plg) results in fibrin-dependent defects in tissue repair. However, repair within the liver is known to be defective in Plg-deficient (Plgo) mice independent of fibrin clearance and appears to be compromised in part by the poor clearance of necrotic cells. Based on these findings, we examined the hepatic transcriptome after injury in search of transcriptional programs that are sensitive to the Plg/fibrinogen system. To this end, we generated biotinylated cRNA pools from livers of Plgo mice and controls before and after a single dose of the hepatotoxin carbon tetrachloride and hybridized them against high-density oligonucleotide arrays. Analysis of the gene expression platform identified an unexpected transcriptional signature within challenged livers of Plgo mice for pancreatic gene products, including trypsinogen-2, amylase-2, elastase-1, elastase-2, and cholesteryl-ester lipase. Validation studies found that this transcriptional program also contained products of the endocrine pancreas (Reg-1 and insulin genes) and the expression of the pancreatic transcription factors p48 and PDX-1. By using a LacZ transgene to trace the cellular source of pancreatic gene expression, we found that PDX-1 was expressed in albumin-positive cells that were morphologically indistinguishable from hepatocytes, and in albumin-negative epithelioid cells within zones of pericentral injury. More detailed studies revealed that the mechanisms of heterotopic gene expression in Plgo mice required fibrin(ogen). Collectively, these data reveal a regulatory role for the hemostatic factors plasmin(ogen) and fibrin(ogen) in cellular plasticity within adult tissues of the digestive system.
Keywords: development, differentiation, regeneration
Tissue morphogenesis and repair require intact growth circuits juxtaposed to the timely remodeling of extracellular matrix. In the mature liver, growth circuits obey hierarchical signals that control cellular differentiation and proliferation (1, 2). Among these signals, transcription factors control cellular proliferation in response to an injury through the control of metabolic homeostasis (3) or as a consequence of growth stimuli from soluble mediators, such as IL-6, TNF-α, keratinocyte growth factor, and others (2). Soluble factors also play a critical role in tissue repair through proteolysis of extracellular substrates. For example, activation of the serine protease plasminogen (Plg) is not required for liver cell proliferation but is essential for the clearance of necrotic cells and the reorganization of the liver lobule after an acute injury (4).
Plg is a ubiquitous zymogen that is proteolytically converted into the active protease plasmin by urokinase- or tissue-type Plg activator. The production of plasmin is thought to contribute significantly to matrix remodeling, the inflammatory response, neurodegeneration, tumor progression, and other processes (5, 6). Although one important target of plasmin-mediated proteolysis is provisional fibrin matrices (5), it is clear that fibrin does not constitute the sole physiologically relevant plasmin substrate. Early evidence of a broader physiological role for the Plg activation system outside fibrinolysis was the finding that plasmin(ogen) supports the reorganization of the hepatic matrix and clearance of necrotic tissue after acute liver injury through a mechanism that is independent of fibrin deposition (4). To better understand the role(s) of plasmin(ogen) in hepatic repair, we carried out a large-scale analysis of liver gene expression at different phases of the repair process in Plg-deficient (Plgo) mice after an acute hepatotoxic injury. We found that Plg deficiency resulted in an entirely unanticipated emergence of cells displaying a pancreatic phenotype, with the expression of pancreatic transcription factors and products of endocrine and exocrine pancreas in a fibrin-dependent fashion. These data reveal a role for plasmin(ogen) and fibrin(ogen) in the regulation of intertissue cellular plasticity among organs of the digestive system in response to an injury.
Methods
Gene-Targeted Mice. Mice with the targeted disruption of Plg (Plgo), fibrinogen (Fibo), or both genes (Plgo/Fibo) were of mixed genetic background (5). PDXlacZko+/- mice were obtained from C. Wright (Vanderbilt University, Nashville TN) and mated with Plgo mice to generate a mouse line with the homozygous inactivation of the Plg gene and heterozygous disruption of the PDX-1 gene by the in-frame insertion of the lacZ minigene (7). All experiments were performed in 1- to 5-month-old mice, pairing littermates to control for all genotypes (Fib+/Plg+, Plgo, Fibo, Plgo/Fibo, Plg+/PDXlacZko+/-, and Plgo/PDXlacZko+/-). Animal protocols were approved by the Institutional Animal Care and Use Committee of the Cincinnati Children's Research Foundation.
Models of Liver Injury and Cell Fractionation. Gene-targeted and control mice were subjected to an acute toxic injury by a single i.p. injection of 0.5 ml of CCl4 (Sigma-Aldrich) per kg of body weight as a 25% (vol/vol) solution in corn oil, examined daily, and killed 2, 7, and 14 days later as described in ref. 4; a separate group of mice was killed before CCl4 injection to serve as controls. When mice were used for liver cell isolation, the portal vein was canulated and perfused with collagenase-containing Hank's buffered salt solution, and the cells were passed through a double layer of gauze as described in ref. 8. After centrifugation at 20 × g for 2 min, parenchymal cells were isolated and kept as a single fraction or treated with pronase to select for cholangiocytes (9), whereas nonparenchymal cells were recovered after additional centrifugation of the supernatant. Phenotypic identification of hepatocyte, cholangiocytes, and nonparenchymal cells was done by quantification of mRNA levels for albumin, cytokeratin-7, and vimentin by real-time PCR (see below). Pancreas and salivary glands were also harvested and immediately frozen in liquid nitrogen for RNA studies or used for protein isolation as described below.
Microarray Studies. Total RNA was isolated from frozen liver samples of Plgo and Plg+ mice before (time 0) and at 2, 7, and 14 days after CCl4 injection using the TRIzol reagent (GIBCO/Life Technologies, Rockville, MD) (10). Equal amounts of RNA from three livers of Plgo or Plg+ mice were pooled at each time point, and biotinylated cRNAs were synthesized for each RNA pool by using 20 μg of total RNA and the SuperScript system (Life Technologies, Grand Island, NY) with poly(dT) primer (10). Each cRNA synthesis reaction was hybridized to the high-density oligonucleotide-based Affymetrix U74Av2 Gene-Chip containing 15,099 gene products with low redundancy. All protocols for chip hybridization, raw and normalized experimental data, bioinformatics approach with statistical analysis, and gene lists are outlined in the MIAME (minimum information about a microarray experiment) guidelines and can be obtained from the authors upon request. In brief, specific hybridization and gene expression were monitored by image analysis of the chip with Affymetrix microarraysuite 5.0. A single platform of gene expression was created with GeneSpring 6.0 (Silicon Genetics, Redwood City, CA) and initially analyzed to identify genes in Plgo livers with levels of expression at least 1.5-fold above Plg+ littermates at each time point using ANOVA and a P < 0.05. We then mined the platform by using the Drawable Gene function of the software to select genes uniquely up-regulated at each time before and after CCl4 injection, with baseline levels at all other time points in Plg+ and Plgo mice. This approach allows for the identification of genes expressed exclusively at single time points and has been successfully used to determine the molecular signatures and predominant physiologic consequences of hepatobiliary obstruction (11).
Identification of Regulatory Motifs. To identify DNA regulatory motifs shared by groups of functionally related genes, we used trafac, an application that surveys for conserved DNA sequences, such as transcription factor-binding sites, between genes (12). In brief, 3 kb of DNA sequence upstream from the 5′ start sites of the genes encoding trypsinogen-2, amylase-2, elastase-1, elastase-2, and cholesteryl-ester lipase were screened for conserved regions by trafac. In this analysis, trafac integrated the conserved sequences identified by repeatmasker, the pipmaker-blastz algorithm, matinspector professional, and match and generated graphical outputs for the entire 3 kb highlighting the putative binding sites and position of homology. Finally, the sites were examined to select regulatory motifs shared by at least three of the genes.
Gene Expression Studies. The expression of individual genes was validated by standard PCR assays using specific primers and stringent cycling conditions (Table 1, which is published as supporting information on the PNAS web site). Quantification of gene expression was determined by real-time PCR using the SYBR green QPCR kit in a Mx4000 thermocycler (Stratagene), with the PCR reaction using 100 ng of cDNA template and the following cycling conditions: 95°C for 10 min, 45 amplification cycles at 95°C for 1 min, annealing for 1 min at specific temperature based on each primer pair (Table 2, which is published as supporting information on the PNAS web site), and extension at 72°C for 1 min.
Cellular Expression Studies. Immunohistochemical analyses of paraffin-embedded liver samples was done with antibodies against amylase (Calbiochem), albumin (Bethyl Laboratories, Montgomery, TX), Fib (4), cytokeratin (DAKO Cytomation, Carpinteria, CA), and PDX-1 (kindly donated by C. Wright). Detection of protein signal was done with species-specific secondary antibodies and the DAB detection system from Vector Laboratories (Burlingame, CA) (4, 13); Texas red-labeled anti-goat antibody (Jackson ImmunoResearch) was used for fluorescence-based immunohistochemical detection of the albumin signal. Sections of pancreas were also included as positive control for specific signals in immunohistochemical assays. X-gal staining was performed in tissues fixed in 4% paraformaldehyde as described in ref. 8.
Amylase Activity Gel. Amylase activity was detected by using starch gel assay as described in ref. 14, which identified pancreas-specific amylase isoform (amylase-2) or salivary isoform (amylase-1). In brief, liver, pancreas, and submaxillary glands were dissected and homogenized in 0.01 M PBS with protease inhibitor mixture (Calbiochem) and centrifuged at 3,300 × g for 5 min. The supernatant was collected and assayed for total protein content by using the Bradford method (Bio-Rad). For each sample, 150 μg of protein was electrophoresed under native gel conditions at 250 V for 6 h, followed by incubation at 37°C for 30-60 min in 2% potato-starch solution (Sigma-Aldrich), two 15-min washes in water, and staining by addition of KI/I2 solution.
Results
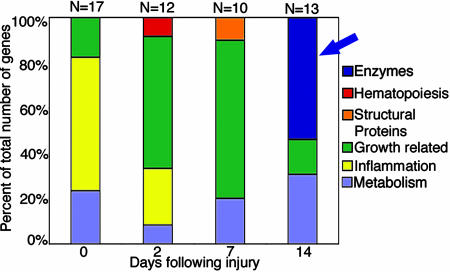
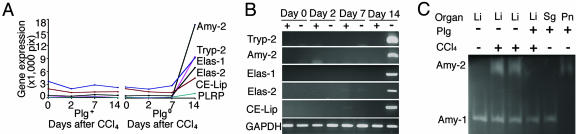
Expression of Genes from Exocrine Pancreas in Plgo Livers. Administration of one dose of the hepatotoxin CCl4 to Plg+ and Plgo mice resulted in a defective liver repair only in Plgo mice, with persistent centrilobular lesions throughout the 14 days, whereas Plg+ mice restored normal lobular appearance by 7 days (Fig. 7, which is published as supporting information on the PNAS web site), as described in ref. 4. To interrogate the hepatic transcriptome for genetic programs that are activated in the context of Plg deficiency, we generated biotinylated cRNA pools from liver samples before and at 2, 7, and 14 days after CCl4 treatment and hybridized them against Affymetrix GeneChips. We then mined the gene expression platform to identify genes that were uniquely up-regulated at specific phases of injury and repair in Plgo livers by at least 1.5-fold above CCl4-treated controls (Plg+ mice). We selected only up-regulated genes to identify molecular signatures of predominant biological processes that are activated in a time-restricted fashion in response to Plg deficiency during liver repair. A total of 52 genes increased exclusively at each phase of hepatic repair (Fig. 1); the complete gene list can be seen in Table 3, which is published as supporting information on the PNAS web site). These genes formed a transcriptional signature of several biological processes that are shared at different phases of injury/repair (such as inflammation, growth control, and metabolism), and a unique expression of pancreatic proteases and hydrolases (trypsinogen-2, amylase-2, lipase-1, lipase-2, and cholesteryl-ester lipase) 14 days after CCl4 injury (Fig. 2 A and B). This transcriptional signature was validated at the protein level by the demonstration of amylase-2 activity in livers of Plgo mice (Fig. 2C) and was the first indication that Plg potentially serves a critical but entirely unanticipated role in controlling a pancreatic phenotype within the hepatic cellular environment.
Fig. 1.
Unique up-regulation of enzymes in Plgo livers. Number of genes overexpressed in Plgo livers is at least 1.5-fold above Plg+ littermates before (day 0) and at 2, 7, and 14 days after CCl4 administration. The number of genes is expressed as a percentage of the total number of genes for each time point, and the genes are grouped based on biological functions. The blue arrow points to the unique up-regulation of a group of genes encoding pancreatic enzymes 14 days after CCl4 administration.
Fig. 2.
Plg deficiency induces the hepatic expression of exocrine pancreatic genes during liver repair. (A and B) The high levels of expression of exocrine pancreatic genes exclusively at 14 days after toxic injury in Plgo livers as determined by the Affymetrix GeneChip (A) or PCR (B). tryp, trypsinogen; amy, amylase; elas, elastase; CE-Lip, cholesterol-ester lipase. (C) A zymographic analysis shows pancreas-specific amylase-2 (Amy-2) activity in livers of Plgo mice 14 days after CCl4 (and in normal pancreas as a positive control), whereas amylase-1 (Amy-1) activity is present in all liver samples and salivary gland tissue. The key indicates the Plg genotypes and whether CCl4 was administered to mice. Li, liver; Sg, salivary gland; Pn, pancreas.
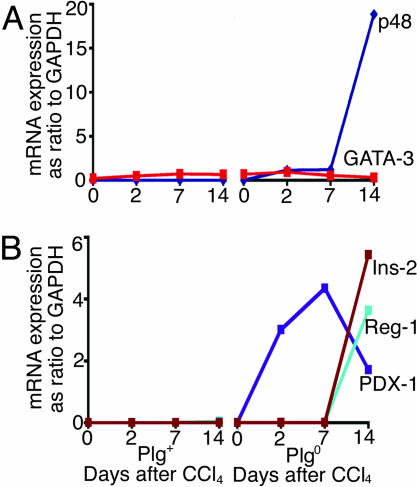
Activation of Pancreas-Specific Transcription Factors in Plgo Livers. To identify potential molecular mechanisms underlying the transcriptional reprogramming, we searched for any shared regulatory sequences within the proximal promoter regions of the five pancreas-enriched genes by using trafac analysis (12). Among the putative transcription factor binding motifs identified, only GATA-3 and p48-PTF-1 binding motifs were present in all five genes (Fig. 8 and Table 4, which are published as supporting information on the PNAS web site). GATA-3 is a member of transcription factors regulating T-cell receptor gene activation (15), whereas p48 is a subunit of the pancreatic transcription factor-1, which is required for the development of the exocrine pancreas (16). Because GATA-3 and p48 were not represented in the gene lists from the microarray, we used real-time PCR to determine the expression of these transcription factors within the liver. Consistent with a targeted transcriptional reprogramming to a pancreatic phenotype in Plgo livers, the expression of p48 increased after CCl4 injury (Fig. 3A), whereas the expression of GATA-3 did not change. To investigate whether such reprogramming encompassed both the exocrine and endocrine cell lineage boundaries, we used the same approach to determine the expression of PDX-1 (transcription factor regulating pancreatic embryogenesis), insulin, and Reg-1 (17-19). Interestingly, although the expression of insulin and Reg-1 was prominent only in Plgo livers and only at 14 days after CCl4, PDX-1 expression increased at 2 days and peaked at day 7 (Fig. 3B). Taken together, these data imply that the loss of Plg in the context of liver repair leads to the expression of transcription factors known to positively regulate the gene expression of exocrine and endocrine pancreas. Interestingly, the coordinate time-restricted expression of pancreatic transcription factors shared the hierarchical profile typical of pancreatic embryogenesis, whereby PDX-1 expression precedes the expression of p48, insulin, and other pancreatic markers (18). Notably, this expression occurred in the context of normal levels of expression of liver-specific genes, such as albumin, prothrombin, and α1-antitrypsin (Fig. 9, which is published as supporting information on the PNAS web site) and was long-lasting in the majority of mice, as demonstrated by the expression of pancreatic genes in five of eight mice 30 days after CCl4 administration (data not shown).
Fig. 3.
Temporal expression of transcription factors and genes of the endocrine pancreas during liver repair. (A) The levels of mRNA expression for p48 and GATA-3 were determined by real-time PCR and show an increase in p48 14 days after CCl4.(B) A similar time-restricted pattern for PDX-1, insulin-2 (Ins-2), and Reg-1.
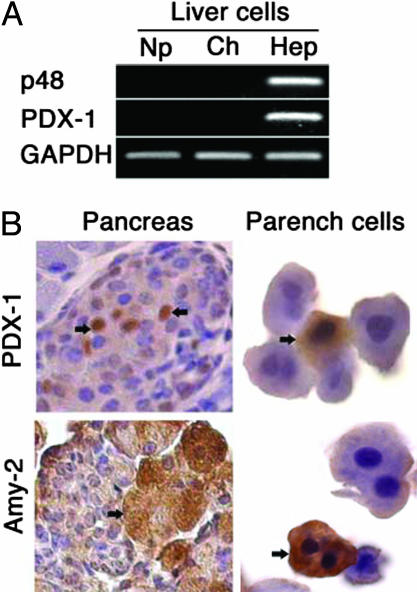
The Pancreatic Phenotype Is Restricted to Liver Parenchymal Cells. To determine which liver cell type expressed pancreas-enriched genes, we isolated populations of parenchymal and nonparenchymal liver cells from Plg+ and Plgo mice 14 days after CCl4 treatment and used real-time PCR to determine gene expression. Among the cell populations enriched for hepatocytes, cholangiocytes, and nonparenchymal cells, only hepatocytes expressed p48 and PDX-1 (Fig. 4A). Immunohistochemical staining for PDX-1 and amylase-2 revealed that these cells were morphologically similar to hepatocytes (Fig. 4B), consistent with the emergence of a cell population within the liver displaying a pancreatic phenotype. The presence of the pancreatic phenotype in Plgo livers was not associated with any histomorphological abnormalities in the pancreas or with notable differences in the pattern of expression of PDX-1 and amylase-2 in the pancreas of Plgo mice and controls 14 days after CCl4 administration (Fig. 10, which is published as supporting information on the PNAS web site).
Fig. 4.
Liver cell expression of pancreatic markers. (A) mRNA expression for PDX-1 and p48 was detected by PCR in isolated hepatocytes (Hep) but not in nonparenchymal cells (Np) or cholangiocytes (Ch). (B) When isolated parenchymal cells are stained with antibodies against PDX-1 or amylase, specific signals are restricted to a subset of cells that share morphological appearance with hepatocytes (brown staining, arrows). Arrows in sections of the pancreas point to signal for PDX-1 in islet cells or for amylase (Amy-2) in acinar cells. (Magnification: Left, ×400; Right, ×1,000.) Parench, parenchymal.
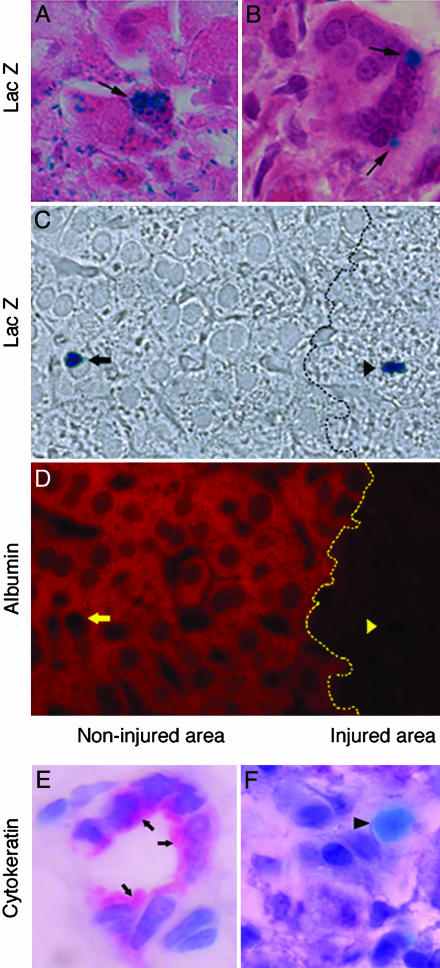
To directly examine the anatomical disposition of these cells within the liver lobule, we histologically flagged PDX-1-expressing cells in Plgo mice through crosses with an available mouse line carrying an in-frame insertion of a lacZ minigene into the PDX-1 locus (PDXlacZko+/-) (7). As expected, hepatotoxic injury of Plg+/PDXlacZko+/- mice did not result in any lacZ expression within the liver after CCl4 (data not shown). However, parallel analyses of the livers collected from Plgo/ PDXlacZko+/- mice revealed the presence of lacZ-expressing cells with morphologic features indistinguishable from hepatocytes or organized in epithelioid structures near the zones of centrilobular injury (Fig. 5 A and B). These cells occurred in a low frequency (<1% of the liver cell population) within noninjured areas or at the margins of centrilobular injury, suggesting that some local cue or stochastic cellular event triggers the expression of pancreatic markers in hepatic cells in Plgo mice. Interestingly, the lacZ-positive cells within the lobule retained the expression of the hepatocyte marker albumin, whereas those in injured zones did not coexpress albumin (Fig. 5 C and D). We found no evidence of a biliary phenotype in lacZ-positive cells, as demonstrated by the lack of cytokeratin staining (Fig. 5 E and F). Collectively, these data indicate that cells with the pancreatic phenotype may share lineage with hepatocytes and may undergo further differentiation and cease expression of hepatocyte markers.
Fig. 5.
Localization of pancreatic cells within the hepatic lobule. (A) X-Gal staining identifies PDX-1 promoter-driven LacZ expression in cells morphologically similar to hepatocytes near the centrilobular injury (arrow). (B) Some of the stained cells also have an epithelioid appearance, are organized in clusters, and are at the transition between injured zones and intact lobule (arrows). (C) LacZ staining in cells of noninjured (arrow) and injured (arrowhead) areas of the liver lobule without background staining. (D) The same section stained with an anti-albumin antibody (red color) depicting albumin-specific signal in LacZ-stained cells only within noninjured areas (yellow arrow). Dotted black and yellow lines indicate the transition between the noninjured area and injured areas. (E and F) Sections of an intrahepatic bile duct containing epithelial cells stained with anti-cytokeratin antibody (E; arrows), whereas LacZ-stained cells (light blue; arrowhead) are negative for cytokeratin (F). (Magnification: E and F, ×1,000.)
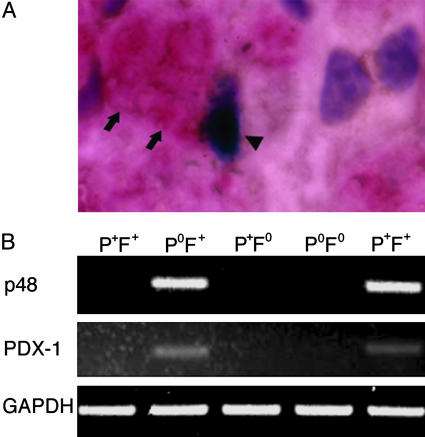
Fibrin Is Required for the Pancreatic Phenotype in the Liver of Plgo Mice. The fibrin-rich extracellular matrix has been shown to regulate the activated phenotype of nonparenchymal hepatic stellate cells (20, 21). To investigate whether fibrin accumulation is associated with the pancreatic phenotype in liver cells, we performed anti-fibrin immunohistochemistry and lacZ staining simultaneously in livers of Plgo mice after CCl4 administration. We found that fibrin staining was juxtaposed to nearly all lacZ-positive cells in livers of Plgo mice (Fig. 6A). Because not all liver cells with pericellular accumulation of fibrin expressed lacZ, we directly explored whether the loss of this physiologically relevant target of plasmin influenced the development of the pancreatic phenotype. To this end, we examined hepatic repair and the differential expression of pancreatic genes in mice with single or combined deficiencies in Plg and Fib (Plgo and Plgo/Fibo). Consistent with our previous report (4), the loss of Plg resulted in a persistent centrilobular injury regardless of the presence or absence of fibrin(ogen) 14 days after CCl4 treatment, whereas control mice and animals lacking fibrin(ogen) alone exhibited complete repair within the same time frame (data not shown). However, although fibrin(ogen) deficiency could not correct the defect in hepatic repair of Plgo mice, the gene expression of exocrine and endocrine pancreas was sensitive to, and entirely dependent on, the presence of Fib (Fig. 6B and Table 5, which is published as supporting information on the PNAS web site). Therefore, fibrin-rich matrices within the injured hepatic microenvironment are mechanistically linked to hepatic cellular plasticity and the activation of a pancreatic transcriptional program in the liver. However, the lack of acquisition of the pancreatic phenotype in Plgo/Fibo mice did not influence the outcome of defective liver repair, as supported by similar morphological appearance and extent of injury in centrilobular areas of Plgo/Fibo mice when compared with Plgo littermates (data not shown). Collectively, these data imply that the hepatic expression of pancreatic markers and of exocrine pancreases, in particular, was triggered by fibrin accumulation in areas of persistent centrilobular injury but was not sufficient to correct the defective repair resulting from the loss of plasmin-mediated proteolysis.
Fig. 6.
Lack of expression of pancreatic genes in mice lacking Plg and Fib. (A) Staining of Fib (arrows) near a LacZ-stained cell (arrowhead) in Plgo liver 14 days after CCl4 injury. (B) mRNA expression for PDX-1 and p48 was detected by PCR in the livers of Plgo mice but is absent in Fibo/Plgo mice and control littermates 14 days after CCl4. The rightmost lane shows expression in the pancreas. Genotypes are shown: P+F+, normal; PoF+, Plg deficiency; P+Fo, Fib deficiency; PoFo, Plgo/Fibo.
Discussion
The unexpected findings of a pancreatic transcriptional program during liver repair in Plgo mice uncover a role for hemostatic factors in the maintenance of cellular phenotype in adult tissues. The spontaneous emergence of such heterotopic phenotype in the liver cells of Plgo mice highlights the potential intertissue plasticity of cells within the digestive system and the potential of components of the hemostatic system to control cellular plasticity. The hepatic cells with a pancreatic expression profile appear to have morphological features of hepatocytes or form clusters of epithelioid cells. Although we have not formally excluded that the cells expressing pancreatic markers emerge from nonparenchymal liver cells (such as macrophages and sinusoidal endothelial cells), several observations favor the view that they are derived, at least in part, from fully differentiated hepatocytes. Specifically, the location of these cells within cords of hepatocytes in the liver lobule, the morphological appearance (including mononucleate and binucleate cells), and the coexpression of albumin in the subset of the cells located within the noninjured portion of the liver lobule all point toward a hepatocyte-related precursor.
The activation in Plgo livers of a transcriptional program that is typical of the primordial endoderm suggests that cells undergoing a hepatic-to-pancreatic switch recapitulate hierarchical signals for pancreatic embryogenesis, with a rise in PDX-1 followed by p48, insulin, and pancreatic enzymes (22). Interestingly, this process occurred without formation of pancreatic islets or acinus, as previously described in a patient with cirrhosis (23), and it did not lead to additional injury to the hepatic lobule, as previously observed in mice overexpressing pancreatic transcription factors (24, 25). One clue regarding the determinants of the hepatic-to-pancreatic switch is that the cells displaying pancreatic products were found exclusively near or within the injured microenvironment. This finding would be consistent with the concept that at least one determinant is a nondiffusible, plasmin-sensitive matrix component that is only present at the sites of injury: Fibrin would be a prime suspect. In this context, the activation of a pancreatic transcriptional program by native hepatic cells may be an inappropriate response to a defect in the repair caused by persistent fibrin within the matrix. This defect appears to ultimately redirect a subset of local hepatic cells toward an alternative genetic program that remains developmentally available. An important interference to this hypothesis is that there may be a persistence of an innate cellular plasticity in some types of fully differentiated cells within the digestive organs throughout adulthood.
The well established impediment in liver repair in Plgo mice may constitute a key regulator of the hepatic-to-pancreatic switch in these animals. This view is supported by the finding that fibrin(ogen) deficiency failed to correct the defective repair in Plgo livers, but it completely eliminated the pancreatic switch. This observation reveals the role of fibrin(ogen) as a potential determinant of the differentiation state of hepatocytes in vivo. Because most mammalian cells appear to maintain the means to engage fibrin-rich matrices (26), these matrices may alter the genetic program in hepatocytes through the engagement of this provisional matrix component or indirectly through soluble or insoluble cues elicited by inflammatory cells in response to persistent fibrin engagement (27). Studies using an in vitro culture system may be helpful in further defining the mechanisms coupling fibrin and fibrinolytic factors to alterations in the genetic program of hepatocytes, Well defined in vitro systems will also be helpful in establishing whether the pancreatic phenotype may be amplified by known soluble mediators of pancreatic function, such as high concentrations of glucose. Fibrin serves as a master regulator of the hepatic genetic program; however, the biological role of fibrin in the control of cell differentiation may be far more general, as supported by previous findings that fibrin controls the growth and differentiation of Schwann cells (28).
The innate intertissue plasticity of the digestive system may have implications regarding potential therapeutic strategies for repopulating a diseased organ with fully functional cells. Several cell lineages have been generated in vivo by transplanted hematopoietic and neural stem cells (29, 30) either by fusion with tissue-specific cells (31-33) or through direct transdifferentiation into mature epithelial cells (34). Similar experimental strategies will be necessary to determine whether the liver cells with pancreatic phenotype identified in our studies can repopulate diseased livers in cell transplantation models, either maintaining their pancreatic program or restoring the liver-specific phenotype of fully differentiated parenchymal cells. In considering effective strategies for tissue repopulation, the identification of fibrin(ogen) as a factor that can modulate cellular plasticity in injured tissues may be useful. If the precise mechanisms linking fibrin(ogen) to cell plasticity could be defined, then protocols might be established whereby fibrin matrices or related factors could be used in tissue engineering or organ regeneration. In this context, detailed studies of hepatic plasticity in Plgo mice may provide a powerful in vivo model to define the components of the hemostatic system that activates alternative genetic programs.
Supplementary Material
Acknowledgments
We thank Drs. William Balistreri, James Wells, and Gail Deutsch for insightful review of the manuscript and experimental support; Dr. Bruce Aronow and Sarah Williams for bioinformatics support; and Dr. Christopher Wright for donation of heterozygous PDXlacZko+/- mice. This work was supported in part by National Institutes of Health Grants DK-55710 (to J.A.B.) and DK-064403 (to the Digestive Disease Research Development Core Center, Cincinnati).
Author contributions: K.S., J.L.D., and J.A.B. designed research; K.S., R.M., G.E.S., and J.A.B. performed research; K.S., R.M., G.E.S., J.L.D., and J.A.B. analyzed data; and K.S., J.L.D., and J.A.B. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Plg, plasminogen; Fib, fibrinogen; Plgo, Plg-deficient; Fibo, Fib-deficient.
References
- 1.Haber, B. A., Mohn, K. L., Diamond, R. H. & Taub, R. (1993) J. Clin. Invest. 91, 1319-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fausto, N. (2001) Liver Transpl. 7, 835-844. [DOI] [PubMed] [Google Scholar]
- 3.Greenbaum, L. E., Li, W., Cressman, D. E., Peng, Y., Ciliberto, G., Poli, V. & Taub, R. (1998) J. Clin. Invest. 102, 996-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bezerra, J. A., Bugge, T. H., Melin-Aldana, H., Sabla, G., Kombrinck, K. W., Witte, D. P. & Degen, J. L. (1999) Proc. Natl. Acad. Sci. USA 96, 15143-15148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bugge, T. H., Kombrinck, K. W., Flick, M. J., Daugherty, C. C., Danton, M. J. & Degen, J. L. (1996) Cell 87, 709-719. [DOI] [PubMed] [Google Scholar]
- 6.Andreasen, P. A., Egelund, R. & Petersen, H. H. (2000) Cell. Mol. Life Sci. 57, 25-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Offield, M. F., Jetton, T. L., Labosky, P. A., Ray, M., Stein, R. W., Magnuson, M. A., Hogan, B. L. & Wright, C. V. (1996) Development (Cambridge, U.K.) 122, 983-995. [DOI] [PubMed] [Google Scholar]
- 8.Yazigi, N. A., Carrick, T. L., Bucuvalas, J. C., Schmidt, C. S., Balistreri, W. F. & Bezerra, J. A. (1997) Transplantation 64, 816-820. [DOI] [PubMed] [Google Scholar]
- 9.Alpini, G., Lenzi, R., Zhai, W. R., Liu, M. H., Slott, P. A., Paronetto, F. & Tavoloni, N. (1989) Gastroenterology 97, 1248-1260. [DOI] [PubMed] [Google Scholar]
- 10.Bezerra, J. A., Tiao, G., Ryckman, F. C., Alonso, M., Sabla, G. E., Sneider, B., Sokol, R. J. & Aronow, B. J. (2002) Lancet 360, 1563-1659. [DOI] [PubMed] [Google Scholar]
- 11.Campbell, K. M., Sabla, G. E. & Bezerra, J. A. (2004) J. Hepatol. 40, 14-23. [DOI] [PubMed] [Google Scholar]
- 12.Jegga, A. G., Sherwood, S. P., Carman, J. W., Pinski, A. T., Phillips, J. L., Pestian, J. P. & Aronow, B. J. (2002) Genome Res. 12, 1408-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pohl, J. F., Melin-Aldana, H., Sabla, G., Degen, J. L. & Bezerra, J. A. (2001) Am. J. Pathol. 159, 2179-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darlington, G. J., Tsai, C. C., Samuelson, L. C., Gumucio, D. L. & Meisler, M. H. (1986) Mol. Cell. Biol. 6, 969-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Labastie, M. C., Bories, D., Chabret, C., Gregoire, J. M., Chretien, S. & Romeo, P. H. (1994) Genomics 21, 1-6. [DOI] [PubMed] [Google Scholar]
- 16.Kawaguchi, Y., Cooper, B., Gannon, M., Ray, M., MacDonald, R. J. & Wright, C. V. (2002) Nat. Genet. 32, 128-134. [DOI] [PubMed] [Google Scholar]
- 17.Akiyama, T., Takasawa, S., Nata, K., Kobayashi, S., Abe, M., Shervani, N. J., Ikeda, T., Nakagawa, K., Unno, M., Matsuno, S. & Okamoto, H. (2001) Proc. Natl. Acad. Sci. USA 98, 48-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guz, Y., Montminy, M. R., Stein, R., Leonard, J., Gamer, L. W., Wright, C. V. & Teitelman, G. (1995) Development (Cambridge, U.K.) 121, 11-18. [DOI] [PubMed] [Google Scholar]
- 19.Fukui, H., Kinoshita, Y., Maekawa, T., Okada, A., Waki, S., Hassan, S., Okamoto, H. & Chiba, T. (1998) Gastroenterology 115, 1483-1493. [DOI] [PubMed] [Google Scholar]
- 20.Ng, V. L., Sabla, G. E., Melin-Aldana, H., Kelley-Loughnane, N., Degen, J. L. & Bezerra, J. A. (2001) J. Hepatol. 35, 781-789. [DOI] [PubMed] [Google Scholar]
- 21.Jang, Y. Y., Collector, M. I., Baylin, S. B., Diehl, A. M. & Sharkis, S. J. (2004) Nat. Cell Biol. 6, 532-539. [DOI] [PubMed] [Google Scholar]
- 22.Wilson, M. E., Scheel, D. & German, M. S. (2003) Mech. Dev. 120, 65-80. [DOI] [PubMed] [Google Scholar]
- 23.Wolf, H. K., Burchette, J. L., Jr., Garcia, J. A. & Michalopoulos, G. (1990) Am. J. Surg. Pathol. 14, 590-595. [DOI] [PubMed] [Google Scholar]
- 24.Ber, I., Shternhall, K., Perl, S., Ohanuna, Z., Goldberg, I., Barshack, I., Benvenisti-Zarum, L., Meivar-Levy, I. & Ferber, S. (2003) J. Biol. Chem. 278, 31950-31957. [DOI] [PubMed] [Google Scholar]
- 25.Horb, M. E., Shen, C. N., Tosh, D. & Slack, J. M. (2003) Curr. Biol. 13, 105-115. [DOI] [PubMed] [Google Scholar]
- 26.Stamatoglou, S. C., Hughes, R. C. & Lindahl, U. (1987) J. Cell Biol. 105, 2417-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flick, M. J., Du, X., Witte, D. P., Jirouskova, M., Soloviev, D. A., Busuttil, S. J., Plow, E. F. & Degen, J. L. (2004) J. Clin. Invest. 113, 1596-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akassoglou, K., Yu, W. M., Akpinar, P. & Strickland, S. (2002) Neuron 33, 861-875. [DOI] [PubMed] [Google Scholar]
- 29.Petersen, B. E., Bowen, W. C., Patrene, K. D., Mars, W. M., Sullivan, A. K., Murase, N., Boggs, S. S., Greenberger, J. S. & Goff, J. P. (1999) Science 284, 1168-1170. [DOI] [PubMed] [Google Scholar]
- 30.Krause, D. S., Theise, N. D., Collector, M. I., Henegariu, O., Hwang, S., Gardner, R., Neutzel, S. & Sharkis, S. J. (2001) Cell 105, 369-377. [DOI] [PubMed] [Google Scholar]
- 31.Alvarez-Dolado, M., Pardal, R., Garcia-Verdugo, J. M., Fike, J. R., Lee, H. O., Pfeffer, K., Lois, C., Morrison, S. J. & Alvarez-Buylla, A. (2003) Nature 425, 968-973. [DOI] [PubMed] [Google Scholar]
- 32.Wang, X., Willenbring, H., Akkari, Y., Torimaru, Y., Foster, M., Al-Dhalimy, M., Lagasse, E., Finegold, M., Olson, S. & Grompe, M. (2003) Nature 422, 897-901. [DOI] [PubMed] [Google Scholar]
- 33.Vassilopoulos, G., Wang, P. R. & Russell, D. W. (2003) Nature 422, 901-904. [DOI] [PubMed] [Google Scholar]
- 34.Harris, R. G., Herzog, E. L., Bruscia, E. M., Grove, J. E., Van Arnam, J. S. & Krause, D. S. (2004) Science 305, 90-93. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.