Abstract
Gene-specific expansion of polyglutamine-encoding CAG repeats can cause neurodegenerative disorders, including Huntington's disease. It is believed that part of the pathological effect of the expanded protein is due to transcriptional dysregulation. Using Drosophila as a model, we show that cAMP-response element-binding protein (CREB) is involved in expanded polyglutamine-induced toxicity. A mutation in the Drosophila homolog of CREB, dCREB2, enhances lethality due to polyglutamine peptides (polyQ), and an additional copy of dCREB2 partially rescues this lethality. Neuronal expression of expanded polyQ attenuates in vivo CRE-mediated transcription of a reporter gene. As reported previously, overexpression of heat-shock protein 70 (Hsp70) rescues polyglutamine-dependent lethality. However, it does not rescue CREB-mediated transcription. The protective effects of CREB and heat-shock protein 70 against polyQ are additive, suggesting that targeting multiple pathways may be effective for treatment of polyglutamine diseases.
Keywords: transcription, polyglutamine diseases, molecular chaperones, PKA
Expansion of polyglutamine-encoding repeats is implicated in the etiology of at least nine neurological disorders (for review, see ref. 1). It is known that long polyglutamine peptides (polyQ) themselves are sufficient to induce neurodegeneration (2), suggesting that the naturally occurring expansions cause disease due to a gain-of-function mechanism. There is evidence that polyQ expansion targets multiple cellular functions, including transcription, proteasomal degradation, molecular chaperones, and axonal transport (for review, see ref. 3).
For some of these diseases, aspects of the pathology are attributable to transcriptional dysregulation. Proteins with expanded polyglutamine stretches can interact with transcriptional cofactors, and transcription is altered in mouse models or patients with polyglutamine expansions (for review, see ref. 4). Transcription cofactors, including cAMP-response element-binding protein-(CREB) binding protein (CBP) and TAFII130, have been found in inclusions in cell culture models or patients of polyglutamine diseases (5-8), and sequestration of these proteins into insoluble aggregates has been suspected to contribute to transcriptional dysregulation. It has been reported that polyQ can inhibit the acetyltransferase activity of CBP (9). Accordingly, inhibition of histone deacetylase activity (9, 10), or overexpression of CBP (8, 11), can reverse the deleterious effects due to expanded polyQ proteins.
CREB is one of the transcription factors that interact with CBP, and it has been implicated in the pathology of polyglutamine diseases. CREB-mediated transcription of reporter genes is attenuated in cultured cells that transiently express polyQ (5-8, 12, 13). Many genes regulated by the cAMP signaling pathway are down-regulated at an early symptomatic stage in cellular and mouse models of Huntington's disease (12, 14). Forskolin stimulation of adenylyl cyclase, as well as overexpression of an activated CREB, ameliorated mutant Huntingtin- (htt) induced phenotypes in a cell culture model (7, 12). Knocking out CREB leads to apoptosis and neurodegeneration in sensory neurons of genetically modified mice (15). Combining the knockout with a null mutation in the closely related family member cAMP-response element modulator (CREM) leads to mice with atrophy of the hippocampus and striatum, a phenotype reminiscent of Huntington's disease patients (16). The levels of cAMP and phosphorylation of CREB decrease in a mouse model with mutant htt that contains 111 glutamine residues (17). However, a recent report shows that phosphorylation of CREB, as well as CRE reporter activity, is enhanced in a transgenic mouse harboring a mutant exon 1 of human htt, suggesting that the role of CREB in polyQ pathology is still unclear (18).
Drosophila models of polyQ diseases have successfully recapitulated some of the pathological features seen in human diseases, including formation of inclusions, cell degeneration, motor dysfunction, and premature death (2, 9, 11, 19-23). In this report, we use the Gal4/upstream activating sequence (UAS) expression system to overexpress polyQ in neurons (2, 9) and test for genetic interaction between polyQ-mediated toxicity and CREB. We demonstrate that a mutation in dCREB2 enhances the lethality of polyQ expressed in the nervous system, whereas an additional copy of dCREB2 partially rescues polyQ-induced lethality. Expression of polyQ in neurons attenuates CRE-mediated reporter activity in vivo. PKA, which increases CREB activity, rescues polyQ-induced lethality as well.
In addition to the restoration of transcriptional activity (4, 9-12), it has been reported that overexpression or activation of molecular chaperones can rescue polyQ-induced toxicity, presumably by increasing solubility of polyQ proteins (21, 24-33). We tested whether the protective effect of the molecular chaperone, heat-shock protein 70 (Hsp70), affected CREB activity. We found that overexpression of Hsp70 rescues polyQ-mediated lethality without recovery of CREB activity, and that the protective effects of CREB and Hsp70 against polyQ are additive.
Materials and Methods
Fly Stocks. gmr-Gal4, elav-Gal4, and S162 flies were obtained from the Bloomington Stock Center, Indiana University. The UAS-polyglutamine lines were gifts from L. M. Thompson and J. L. Marsh (2, 9). A genomic fragment carrying the dCREB2 region was cloned and injected to establish the gdCREB fly lines. A stop codon was engineered into the coding region of the dCREB2 genomic fragment, producing the gdCREB stop transgene. The R4F CA-MEK1 (ΔN3/S218E/S222D) cDNA (34), a gift from N. G. Ahn, was subcloned and used to establish hs-MEK transgenic flies [hs-MEK, the constitutively active form of mitogen-activated protein kinase kinase (MEK) under the control of the heat-shock promoter]. The transgenic flies carrying the CRE-luciferase reporter (CRE-Luc) transgene have been described (35). Transgenic flies carrying the UAS-hsp70 transgene were a gift from N. M. Bonini (University of Pennsylvania, Philadelphia) (27), and the hs-PKA* flies (the constitutively active form of PKA) were from G. Struhl (Columbia University, New York) (36).
Survival Assay. Fies carrying an UAS transgene in trans to a balancer chromosome were crossed to flies carrying the GAL4 transgene. The flies that emerged were collected, and the number of flies with each genotype was counted. The survival rate was calculated by dividing the number of polyQ-expressing flies (flies carrying both the Gal4 and UAS chromosomes, experimental group) by the number that do not express polyQ (control group) and multiplying by 100. More than 200 control flies for each genotype were counted. Experiments were repeated three times, and representative data are shown. The genotypes for all crosses are provided in Supporting Text, which is published as supporting information on the PNAS web site. For analysis, we used the χ2 test, which approximates how observed data relate to the expected outcome of the experiment. The observed data are the number of flies that survive to the adult stage, and the expected outcome is predicted by the number of flies with balancer and Mendelian ratio (37).
Antibodies. Anti-dCREB2 monoclonal and Ser-231 phosphospecific anti-pCREB rabbit polyclonal antibodies have been described (38).
Luciferase Assay. Flies were raised in a 12:12 light/dark cycle at 25°C. For in vitro assays (Fig. 2 A and B), 5-10 fly heads carrying CRE-Luc were homogenized and analyzed by using the Luciferase Assay System (Promega). Luciferase signals were normalized to total protein concentration. Samples were measured in triplicate, and two independent determinations were made. In vivo luciferase activity was recorded from behaving flies as described (35). Briefly, flies carrying CRE-Luc were loaded in 96-well plates filled with agar containing sucrose and luciferin (Biosynth, Basel), covered with PCR caps and clear plastic covers (Topseal, PerkinElmer) that had holes punched in them. Activity was measured by using a Topcount microplate scintillation and luminescence counter (PerkinElmer) under a 12:12 light/dark cycle. For heat-shock induction of MEK, flies were raised at 22°C and received heat shock at 35°C for 30 min per day for 3 days before assaying for luciferase activity at 25°C. Averages from >20 flies are shown.
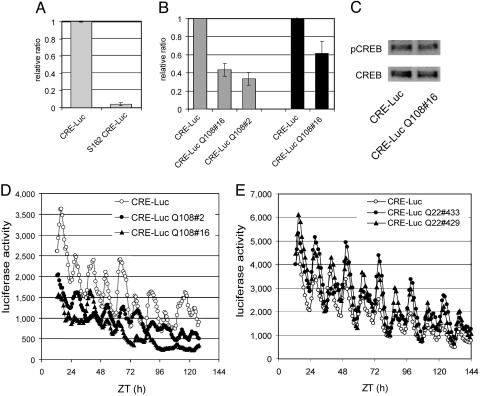
Fig. 2.
Expanded polyQ peptides attenuate CREB-mediated transcription. (A) The CRE-Luc activity in wild-type flies is set to 1 for A and B, and mutants are normalized to this value. CRE-Luc in S162 hemizygous flies (S162 CRE-Luc) is lower than in control (CRE-Luc) flies (n = 5). (B) Expanded polyQ dampens CRE-Luc activity. CRE-Luc activity in flies expressing Q108 in the nervous system (using elav-GAL4 driver, gray bars) or in the eyes (using gmr-GAL4, black bars) is lower than in wild-type (CRE-Luc) flies. Flies were collected at Zeitgeber time = 2.5. Two independent lines carrying the UAS-polyQ108 transgene (#16 and #2) behaved identically. (C) PolyQ does not affect Ser-231 phosphorylation. Western blot of aliquots of the sample used in B probed with anti-dCREB2 (CREB) or anti-dCREB2-P, a phosphospecific antibody that recognizes phosphorylated Ser 231(pCREB). (D) CRE-Luc recording from control (CRE-Luc, open circles) and two independent lines with Q108 expression in neurons (CRE-Luc Q108#2, filled circle, and CRE-Luc Q108#16, filled triangles). (E) Q22 expression does not reduce CRE-Luc activity. CRE-Luc signal from control (CRE-Luc, open circle) and Q22-expressing flies (CRE-Luc Q22#433, filled circles, and CRE-Luc Q22#429, filled triangles).
Climbing Assay. The climbing assay was performed as described (39), with slight modifications. Twenty flies were placed in a plastic vial and gently tapped to the bottom. The number of flies that reached the top of the vials within 18 sec was counted. The mean of 20 trials with SEM is shown. The experiments were repeated independently three times, and similar results were obtained for each replicate.
Results
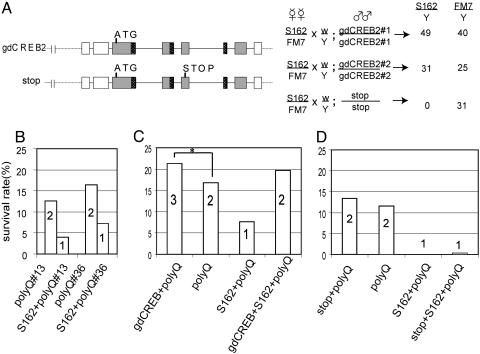
The Gene Dosage of dCREB2 Modifies PolyQ-Induced Lethality. S162 is an X-linked, homozygous lethal mutation in the Drosophila dCREB2 gene (35). This mutation is recessive, because the heterozygous mutation does not affect the survival rate (see below). We confirmed that S162 is a mutation in the dCREB2 gene by using genomic rescue. A transgenic copy of the dCREB2 genomic fragment (gdCREB), but not one engineered to contain a stop codon (stop), rescues viability (Fig. 1A).
Fig. 1.
Genetic interaction between CREB and expanded polyQ. (A) S162 is a mutation in dCREB2. (Left) Cartoon of a 9-kb dCREB2 genomic rescue fragment, showing the coding (gray), alternatively spliced (black), noncoding (white) exons, the translation start (ATG) and engineered stop codon (stop), and the 5′ and 3′ regions. (Right) Crossing S162/FM7 females to transgenic males homozygous for two independent gdCREB insertions (gdCREB#1 and gdCREB#2) but not to those containing the gdCREBstop transgene (stop) results in rescue of S162 males. The number of progeny for each genotype is shown. (B) S162 increases polyQ-mediated lethality. The gene dosage of dCREB2 is indicated in the bars. The percent of survivors is calculated by dividing the number of polyQ progeny flies by the number of expected progeny of each genotype. Two independent UAS-polyQ48 lines (polyQ#13 and polyQ#36) behaved identically. (C) PolyQ-mediated lethality is sensitive to dCREB2 gene dosage. Survival rates of flies containing one (S162+polyQ), two (polyQ, or gdCREB+S162+polyQ), or three copies of genomic dCREB2 (gdCREB+polyQ). dCREB2 partially rescues lethality (significant difference between gdCREB+polyQ and polyQ, P < 0.05, χ2 value 4.19, asterisk). Significant differences between S162+polyQ and polyQ (P < 0.00001, χ2 value 27.3) or S162+polyQ and gdCREB+S162+polyQ (P < 0.00001, χ2 value 46.5) (D) A genomic fragment of dCREB2 with a stop codon (stop) does not alter the survival rate of polyQ-mediated lethality. The survival of polyQ flies was not different from that of stop+polyQ flies (P > 0.05, χ2 value 1.09). The stop+S162+polyQ flies did not survive, similar to the S162+polyQ flies, but unlike flies with two functional copies (C, gdCREB+S162+polyQ).
In the fly model for polyQ-mediated toxicity, various length polyQ (Q22, Q48, and Q108) can be placed under the control of the UAS promoter sequence (2, 9). Elav-Gal4-restricted expression of Q48 or Q108, but not Q22, in postmitotic neurons results in partial lethality (9). We find that the survival rate of flies expressing Q48 in the nervous system is sensitive to the gene dosage of the dCREB2 gene. S162 heterozygous flies show twice the polyQ-mediated lethality as wild-type flies (Fig. 1B). A single copy of S162 itself does not cause any lethality (S162/+ divided by FM7/+ yields 102.8%). This effect is seen with two independent insertions of the polyQ transgene, making it unlikely that the increased lethality is due to the insertion site of either transgene (Fig. 1B). The S162 heterozygous mutation also enhances Q108-mediated lethality (data not shown). This increase in lethality requires the toxic form of polyQ, because Q22 expressed in neurons does not show any increase in toxicity when placed in a heterozygous S162 background (101% for both wild-type and heterozygous strains). This result demonstrates that S162's effect on lethality is specific to toxicity induced through expanded polyQ.
The enhancement of polyglutamine-induced lethality seen in heterozygous S162+poly Q flies is suppressed when the gdCREB transgene, but not the stop transgene, is added back (Fig. 1 C and D). This indicates that the enhancement of lethality due to expanded polyQ is attributable to a loss of dCREB2 function. The gdCREB transgene, when present as a third genomic copy, also partially, but significantly, ameliorates the lethality of polyQ flies (Fig. 1C). Therefore, dCREB2 is sufficient to overcome a significant fraction of polyQ-mediated effects because, depending upon its copy number, it can increase or decrease toxicity.
PolyQ Attenuate CRE-Luc Activity in Vivo. CREB-dependent transcription in vivo can be monitored by using transgenic flies that contain a CRE-Luc (35). Hemizygous S162 “escaper” flies show a dramatic decrease in luciferase activity (Fig. 2A), confirming dCREB2 responsiveness of the reporter (35). When expanded polyQ is expressed pan-neuronally (using an elav-Gal4 driver) or in the eye (using gmr-Gal4), luciferase activity measured biochemically also decreases when compared with control flies (Fig. 2B).
As described (35), dCREB2 activity oscillates in a circadian manner. Luciferase activity in flies with polyQ108 expression still cycles but is lower at all time points (Fig. 2D). The short polyQ (Q22), which does not cause toxicity, also does not attenuate CREB activity (Fig. 2E). Therefore, the attenuation of CRE-Luc activity depends on the length of the polyQ tract that is overexpressed and correlates with toxicity effects of the peptides. However, the reduction of CRE-Luc activity is not due to extreme loss of neurons, because there is little change in the protein level of dCREB2 (Fig. 2C) or in the gross morphology of polyQ-expressing heads (data not shown). PolyQ expression also does not significantly affect phosphorylation at dCREB2 Ser-231, which is the equivalent residue to mammalian Ser-133 (Fig. 2C). The expression of polyQ22 actually increases CRE-Luc activity, perhaps because glutamine tracts themselves can enhance transcription (40).
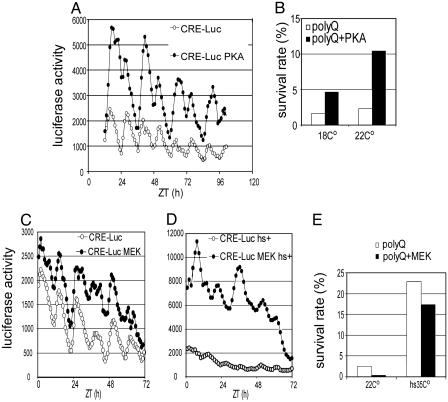
PKA Increases CREB Activity and Rescues PolyQ-Induced Lethality. CREB-responsive transcription is involved in various cellular responses to different stimuli. It is believed that each individual stimulus activates CREB and results in the transcription of a different, but perhaps overlapping, subset of “downstream” genes. The specificity of the target genes is thought to be at least partially due to the details of the upstream pathways that activate CREB (for review, see ref. 41). Therefore, we tested whether manipulating signaling pathways that increase CREB activity could rescue polyQ-induced toxicity. Transgenic flies carrying hs-PKA* (36) show increased CREB activity in vivo at 25°C (Fig. 3A). hs-PKA* increases the survival rate of flies that express expanded polyQ in the nervous system at 18°C and 22°C (Fig. 3B). The increase in survival rate with hs-PKA* is larger at 22°C than at 18°C, suggesting that this effect depends upon the level of PKA* expression. A single copy of hs-PKA* itself does not affect survival rate at these conditions (the number of hs-PKA*/+ divided by that of CyO/+ yields 97.9% at 18°C and 109.9% at 22°C).
Fig. 3.
Effect of PKA and MEK on CRE-luciferase activity and polyQ-induced lethality. (A) Luciferase activity from control (CRE-Luc, open circles) and transgenic flies with hs-PKA* (CRE-Luc PKA, filled circles). (B) Survival rates for flies with (polyQ+PKA, filled bars) or without hs-PKA* (polyQ, open bars), raised at 18°C or 22°C. (C) Luciferase signal from control flies (CRE-Luc, open circles) and that from flies with hs-MEK (CRE-Luc MEK, filled circles). (D) Luciferase signal from the control (CRE-Luc hs+, open circles) or hs-MEK (CRE-Luc MEK hs+, filled circles) flies that were both exposed to heat shock. (E) Survival rates for the flies with (polyQ+MEK, filled bars) or without (polyQ, open bars) the hs-MEK transgene when they were raised at 22°C (22°C) or raised at 22°C and heat-shocked at 35°C for 30 min every day (hs35°C).
We tested the effect of another kinase, MEK, using transgenic flies carrying hs-MEK. hs-MEK flies, when raised at 22°C (Fig. 3C) or when heat-induced (Fig. 3D) have higher CRE-Luc activity than control flies. However, hs-MEK does not rescue polyQ-induced lethality in flies raised at 22°C or those that receive heat shock (Fig. 3E), even though the increase in CRE-Luc activity after hs-MEK induction is greater than that achievable with PKA (compare Fig. 3 C and D). The survival rate of polyQ flies that receive heat shock is higher for all genotypes, presumably due to the ability of heat shock itself to rescue polyQ effects (see below). A single copy of hs-MEK itself does not affect survival rate at these conditions [the number of hs-MEK/+ divided by that of CyO/+ yields 100.0% for 22°C and 92.2% for hs35°C, no significant decrease (P > 0.05)]. These data suggest that certain, but not all, signaling pathways that activate CREB can rescue polyQ toxicity.
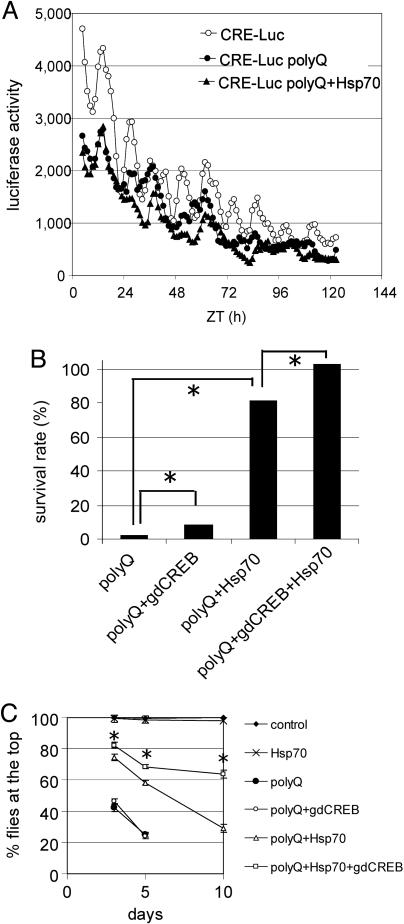
Overexpression of Hsp70 Does Not Rescue PolyQ-Mediated Effects on CREB Activity. Overexpression of heat-shock proteins can rescue polyQ-induced degeneration in cellular, mouse, and Drosophila models (20, 24-27, 29, 30, 42-44). However, it is unknown whether this protective effect of heat-shock proteins involves reversal of transcriptional defects. We tested whether Hsp70-mediated rescue affected CREB activity in flies with expanded polyQ expression. CRE-Luc activity remains depressed in flies that overexpress polyQ and Hsp70 (Fig. 4A). However, overexpression of Hsp70 greatly suppresses polyQ-mediated toxicity (Fig. 4B, compare polyQ and polyQ+Hsp70), as reported (27). Hsp70 overexpression itself does not change CRE-Luc activity (Fig. 5, which is published as supporting information on the PNAS web site).
Fig. 4.
CREB and Hsp70 additively suppress polyQ-induced toxicity. (A) Overexpression of Hsp70 does not rescue CRE-Luc activity in polyQ-expressing flies. Luciferase activity from control flies (CRE-Luc, open circles), flies with Q108 expression (CRE-Luc polyQ, filled circles), and flies with Hsp70 and Q108 expression in neurons (CRE-Luc polyQ+Hsp70, filled triangles). (B) The protective effect of Hsp70 and dCREB2 against Q108-mediated lethality are additive. The survival rates for the flies with polyQ only, polyQ plus an extra copy of dCREB2 (polyQ+gdCREB), polyQ plus Hsp70 (polyQ+Hsp70), and polyQ plus both gdCREB and Hsp70 (polyQ+gdCREB+Hsp70) are shown. There is a significant difference between polyQ and polyQ+gdCREB, polyQ and polyQ+Hsp70, and polyQ+Hsp70 and polyQ+gdCREB+Hsp70 [χ2 value 62.36, 15.90, and 13.41, respectively; P < 0.0005 (asterisks)]. (C) gdCREB and Hsp70 additively suppress polyQ-mediated defects in the climbing assay. The climbing assay is shown for wild-type control flies and flies that overexpress Hsp70, polyQ, polyQ+gdCREB, polyQ+Hsp70, or polyQ+Hsp70+gdCREB. Asterisks indicate significant differences between polyQ+Hsp70 and polyQ+Hsp70+gdCREB (P < 0.05 with Student's t test).
The Hsp70- and CREB-mediated rescue of polyQ-induced lethality is additive (Fig. 4B). Although the CREB-mediated rescue of lethality is quantitatively much smaller than that of Hsp70, it is statistically significant (Fig. 4B, polyQ vs. polyQ+gdCREB; χ2 value of 62.36, P < 0.000005). When an additional copy of gdCREB was added to flies containing polyQ+Hsp70, there was further significant rescue (polyQ+Hsp70 vs. polyQ+gdCREB+Hsp70; χ2 value 13.4, P < 0.0005). The combined rescue achieved with gdCREB+Hsp70 is greater than the sum of their individual effects, suggesting that the effect of gdCREB and Hsp70 may be synergistic.
Motor defects are commonly seen in Huntington's disease. When polyQ-containing transgenic flies are assayed in a simple locomotor assay, the transgenic flies show a severe decrease in activity (Fig. 4C). In this assay, the gdCREB transgene does not rescue polyQ-mediated locomotor dysfunction, but overexpressed Hsp70 is able to partially rescue. However, similar to the rescue of lethality, gdCREB and Hsp70 additively rescue when cooverexpressed (Fig. 4C, asterisks), and this suppression is more dramatic as the flies age. Collectively, these results demonstrate that overexpression of Hsp70 is not sufficient to reverse decreases in dCREB2 activity, and that Hsp70 and dCREB2 can additively rescue polyQ-induced toxicity.
Discussion
Accumulating evidence indicates that transcriptional dysregulation is involved in the pathogenesis of polyglutamine diseases (for review, see ref. 4). Here, we have shown that CREB is involved in polyQ-mediated toxicity in Drosophila neurons. A heterozygous mutation in the dCREB2 gene decreases the survival rate of flies when expanded polyQ are expressed. An additional genomic copy of dCREB2 slightly but significantly rescues this toxicity. Expression of the expanded polyQ-containing peptides in neurons attenuates CREB-responsive transcription measured using an in vivo reporter. Taken together, these results clearly demonstrate that part of the polyQ-induced lethality can be attributed to inhibition of CREB-mediated transcription.
A number of reports have shown that expression of polyQ in cell culture models attenuates CREB activity (5, 6, 8, 11-13, 45), and a knock-in of 109 CAG repeats in mouse htt (HdhQ111) has been reported to interfere with phosphorylation of CREB and the levels of brain-derived neurotrophic factor mRNA in the striatum (17). Our results are consistent with these reports. Recently, Obrietan and Hoyt (18) reported that phosphorylation of CREB and CRE-reporter activity was enhanced in mice carrying a transgene that codes for an exon 1 of htt with ≈150 glutamine repeat (R6/2 mice). One of the possible explanations for these conflicting results may be that CREB is activated by secondary cellular responses against polyQ toxicity. Various cellular responses, including ischemia (46, 47), oxidative stress (48), or excitotoxicity (49), can activate CREB. In the R6/2 mouse, CREB activity was tested at a relatively late stage (11 weeks) in the degenerative process. Another possibility is that the increase in CREB activity is due to overexpression of htt exon 1 residues that flank the polyQ stretch. Finally, it is possible that the exact protein context that contains the polyQ stretch may contribute to the observed differences.
In cellular and mouse models of Huntington's disease, the cAMP-PKA pathway and the phosphorylation on CREB of Ser-133 is decreased (12, 17). In the fly model described here, there was not a significant change in the phosphorylation level of Ser-231, the fly equivalent to Ser-133 (Fig. 2C). This could be due to differences in the regulatory mechanisms controlling mammalian and fly CREBs. In contrast to mammalian CREB whose activity is mostly correlated with the phosphorylation level of Ser-133, a large portion of dCREB2 exists with Ser-231 in a phosphorylated state, suggesting that other mechanisms must be important in regulating its activity (38). Alternatively, polyQ in this fly model is overexpressed as a peptide, whereas other studies have used polyQ runs in the context of htt protein residues surrounding the expansion. Because protein context plays an important role in the toxicity of polyQ protein (50), and it is likely there are protein context-dependent and independent changes in transcriptional profile in various polyQ diseases (51, 52), it is important to investigate the effect of each protein context on dysregulation of CREB-mediated transcription.
It is believed that polyQ can exist in several physical states, including as monomeric random coils, an oligomeric β sheet form and an SDS-insoluble large aggregate of amyloid fibrils (for review, ref. 53). Overexpression of Hsp70 has been reported to modify the conformation of polyQ proteins before aggregation or to increase the solubility of aggregates (54-59). Transcriptional cofactors have been reported to interact with expanded polyQ (9, 14) or can be sequestered into inclusions (5-8). Recently, Schaffar et al. (60) reported that an htt fragment with expanded polyQ undergoes a conformational change before oligomerization and inhibition of TATA box-binding protein to bind DNA. In vitro, Hsp70/40 is able to reverse this inhibition (60). We found that Hsp70 overexpression does not reverse polyQ inhibition of CREB-mediated transcription (Fig. 4A). One possible interpretation of these results is that there is a pool of toxic polyQ that does not interact with Hsp70. Alternatively, Hsp70 interaction with polyQ may not be sufficient to restore CREB's transcriptional activity.
Conclusion
We have demonstrated that CREB can partially reverse polyQ-mediated toxicity, and that the protective effects of CREB and Hsp70 are additive. Targeting multiple pathways may be required to cure polyglutamine diseases. Hsp70 overexpression has modest effects in several animal models of polyglutamine disease (29, 30, 42-44, 59), and dysregulation of CREB-mediated transcription might contribute to the remaining dysfunction. Recently, Agrawal et al. (61) showed that geladanamycin, an enhancer of Hsp70 activity, and an histone deacetylase inhibitor additively rescue toxicity induced by mutant htt exon 1 in Drosophila, supporting our conclusion. It is reasonable that pharmacological enhancement of CREB activity through specific pathway(s), or augmentation of CREB downstream genes, might ameliorate the dysfunctional effects of polyQ expansion. Drosophila may be a useful system to search for such modifiers, because many functions of CREB, including memory formation and circadian rhythms, are conserved in flies (35, 62, 63).
Supplementary Material
Acknowledgments
We thank Drs. J. L. Marsh (University of California, Irvine) and L. M. Thompson (University of California, Irvine) for the UAS-polyQ fly lines, Dr. N. M. Bonini (University of Pennsylvania, Philadelphia) for UAS-Hsp70 transgenic flies, Dr. G. Struhl (Columbia University, New York) for hs-PKA* transgenic flies, and Dr. N. G. Ahn (University of Colorado, Boulder) for R4F CA-MEK1 (ΔN3/S218E/S222D) cDNA. We thank Sumei Lu and Dr. Joan C. Hendricks for essential help and discussion. We thank Drs. J. Dubnau, C. Margulies, A. Sehgal, T. Tully, and Y. Zhong for comments. K.I.-A. is supported by a long-term fellowship of the Human Frontier Science Program. K.I. is supported by Alzheimer's Association Grant NIRG-03-5239. This work was supported by funding from the National Institutes of Health (MH67774, to J.C.P.Y.).
Author contributions: K.I.-A. and K.I. designed research; K.I.-A. performed research; E.A.D. contributed Fig. 1A; P.W. and E.A.D. contributed new reagents; K.I.-A. and J.C.P.Y. analyzed data; and K.I.-A., K.I., and J.C.P.Y. wrote the paper.
Abbreviations: CREB, cAMP-response element-binding protein; CBP, CREB-binding protein; htt, Huntingtin; polyQ, polyglutamine peptide; UAS, upstream activating sequence; CRE-Luc, CRE-luciferase reporter; hs-PKA*, constitutively active form of PKA; MEK, mitogen-activated protein kinase kinase; hs-MEK, constitutively active form of MEK under the control of the heat-shock promoter.
References
- 1.Zoghbi, H. Y. & Orr, H. T. (2000) Annu. Rev. Neurosci. 23, 217-247. [DOI] [PubMed] [Google Scholar]
- 2.Marsh, J. L., Walker, H., Theisen, H., Zhu, Y. Z., Fielder, T., Purcell, J. & Thompson, L. M. (2000) Hum. Mol. Genet. 9, 13-25. [DOI] [PubMed] [Google Scholar]
- 3.Li, S.-H. & Li, X.-J. (2004) Trends Genet. 20, 146-154. [DOI] [PubMed] [Google Scholar]
- 4.Sugars, K. L. & Rubinsztein, D. C. (2003) Trends Genet. 19, 233-238. [DOI] [PubMed] [Google Scholar]
- 5.Shimohata, T., Nakajima, T., Yamada, M., Uchida, C., Onodera, O., Naruse, S., Kimura, T., Koide, R., Nozaki, K., Sano, Y., et al. (2000) Nat. Genet. 26, 29-36. [DOI] [PubMed] [Google Scholar]
- 6.Nucifora, F. C., Jr., Sasaki, M., Peters, M. F., Huang, H., Cooper, J. K., Yamada, M., Takahashi, H., Tsuji, S., Troncoso, J., Dawson, V. L., et al. (2001) Science 291, 2423-2428. [DOI] [PubMed] [Google Scholar]
- 7.Sugars, K. L., Brown, R., Cook, L. J., Swartz, J. & Rubinsztein, D. C. (2004) J. Biol. Chem. 279, 4988-4999. [DOI] [PubMed] [Google Scholar]
- 8.McCampbell, A., Taylor, J. P., Taye, A. A., Robitschek, J., Li, M., Walcott, J., Merry, D., Chai, Y., Paulson, H., Sobue, G., et al. (2000) Hum. Mol. Genet. 9, 2197-2202. [DOI] [PubMed] [Google Scholar]
- 9.Steffan, J. S., Bodai, L., Pallos, J., Poelman, M., McCampbell, A., Apostol, B. L., Kazantsev, A., Schmidt, E., Zhu, Y. Z., Greenwald, M., et al. (2001) Nature 413, 739-743. [DOI] [PubMed] [Google Scholar]
- 10.Hockly, E., Richon, V. M., Woodman, B., Smith, D. L., Zhou, X., Rosa, E., Sathasivam, K., Ghazi-Noori, S., Mahal, A., Lowden, P. A. S., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 2041-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor, J. P., Taye, A. A., Campbell, C., Kazemi-Esfarjani, P., Fischbeck, K. H. & Min, K. T. (2003) Genes Dev. 17, 1463-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wyttenbach, A., Swartz, J., Kita, H., Thykjaer, T., Carmichael, J., Bradley, J., Brown, R., Maxwell, M., Schapira, A., Orntoft, T. F., et al. (2001) Hum. Mol. Genet. 10, 1829-1845. [DOI] [PubMed] [Google Scholar]
- 13.Dunah, A. W., Jeong, H., Griffin, A., Kim, Y. M., Standaert, D. G., Hersch, S. M., Mouradian, M. M., Young, A. B., Tanese, N. & Krainc, D. (2002) Science 296, 2238-2243. [DOI] [PubMed] [Google Scholar]
- 14.Luthi-Carter, R., Strand, A., Peters, N. L., Solano, S. M., Hollingsworth, Z. R., Menon, A. S., Frey, A. S., Spektor, B. S., Penney, E. B., Schilling, G., et al. (2000) Hum. Mol. Genet. 9, 1259-1271. [DOI] [PubMed] [Google Scholar]
- 15.Lonze, B. E., Riccio, A., Cohen, S. & Ginty, D. D. (2002) Neuron 34, 371-385. [DOI] [PubMed] [Google Scholar]
- 16.Mantamadiotis, T., Lemberger, T., Bleckmann, S. C., Kern, H., Kretz, O., Martin Villalba, A., Tronche, F., Kellendonk, C., Gau, D., Kapfhammer, J., et al. (2002) Nat. Genet. 31, 47-54. [DOI] [PubMed] [Google Scholar]
- 17.Gines, S., Seong, I. S., Fossale, E., Ivanova, E., Trettel, F., Gusella, J. F., Wheeler, V. C., Persichetti, F. & MacDonald, M. E. (2003) Hum. Mol. Genet. 12, 497-508. [DOI] [PubMed] [Google Scholar]
- 18.Obrietan, K. & Hoyt, K. R. (2004) J. Neurosci. 24, 791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson, G. R., Salecker, I., Dong, X., Yao, X., Arnheim, N., Faber, P. W., MacDonald, M. E. & Zipursky, S. L. (1998) Neuron 21, 633-642. [DOI] [PubMed] [Google Scholar]
- 20.Kazemi-Esfarjani, P. & Benzer, S. (2000) Science 287, 1837-1840. [DOI] [PubMed] [Google Scholar]
- 21.Fernandez-Funez, P., Nino-Rosales, M. L., de Gouyon, B., She, W. C., Luchak, J. M., Martinez, P., Turiegano, E., Benito, J., Capovilla, M., Skinner, P. J., et al. (2000) Nature 408, 101-106. [DOI] [PubMed] [Google Scholar]
- 22.Warrick, J. M., Paulson, H. L., Gray-Board, G. L., Bui, Q. T., Fischbeck, K. H., Pittman, R. N. & Bonini, N. M. (1998) Cell 93, 939-949. [DOI] [PubMed] [Google Scholar]
- 23.Takeyama, K., Ito, S., Yamamoto, A., Tanimoto, H., Furutani, T., Kanuka, H., Miura, M., Tabata, T. & Kato, S. (2002) Neuron 35, 855-864. [DOI] [PubMed] [Google Scholar]
- 24.Chai, Y., Koppenhafer, S. L., Bonini, N. M. & Paulson, H. L. (1999) J. Neurosci. 19, 10338-10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cummings, C. J., Mancini, M. A., Antalffy, B., DeFranco, D. B., Orr, H. T. & Zoghbi, H. Y. (1998) Nat. Genet. 19, 148-154. [DOI] [PubMed] [Google Scholar]
- 26.Stenoien, D. L., Cummings, C. J., Adams, H. P., Mancini, M. G., Patel, K., DeMartino, G. N., Marcelli, M., Weigel, N. L. & Mancini, M. A. (1999) Hum. Mol. Genet. 8, 731-741. [DOI] [PubMed] [Google Scholar]
- 27.Warrick, J. M., Chan, H. Y., Gray-Board, G. L., Chai, Y., Paulson, H. L. & Bonini, N. M. (1999) Nat. Genet. 23, 425-428. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto, A., Lucas, J. J. & Hen, R. (2000) Cell 101, 57-66. [DOI] [PubMed] [Google Scholar]
- 29.Adachi, H., Katsuno, M., Minamiyama, M., Sang, C., Pagoulatos, G., Angelidis, C., Kusakabe, M., Yoshiki, A., Kobayashi, Y., Doyu, M., et al. (2003) J. Neurosci. 23, 2203-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cummings, C. J., Sun, Y., Opal, P., Antalffy, B., Mestril, R., Orr, H. T., Dillmann, W. H. & Zoghbi, H. Y. (2001) Hum. Mol. Genet. 10, 1511-1518. [DOI] [PubMed] [Google Scholar]
- 31.Chan, H. Y., Warrick, J. M., Andriola, I., Merry, D. & Bonini, N. M. (2002) Hum. Mol. Genet. 11, 2895-2904. [DOI] [PubMed] [Google Scholar]
- 32.Auluck, P. K. & Bonini, N. M. (2002) Nat. Med. 8, 1185-1186. [DOI] [PubMed] [Google Scholar]
- 33.Sittler, A., Lurz, R., Lueder, G., Priller, J., Hayer-Hartl, M. K., Hartl, F. U., Lehrach, H. & Wanker, E. E. (2001) Hum. Mol. Genet. 10, 1307-1315. [DOI] [PubMed] [Google Scholar]
- 34.Mansour, S. J., Matten, W. T., Hermann, A. S., Candia, J. M., Rong, S., Fukasawa, K., Vande Woude, G. F. & Ahn, N. G. (1994) Science 265, 966-970. [DOI] [PubMed] [Google Scholar]
- 35.Belvin, M. P., Zhou, H. & Yin, J. C. (1999) Neuron 22, 777-787. [DOI] [PubMed] [Google Scholar]
- 36.Jiang, J. & Struhl, G. (1995) Cell 80, 563-572. [DOI] [PubMed] [Google Scholar]
- 37.Sokal, R. R. & Rohlf, F. J. (1995) Biometry: The Principles and Practice of Statistics in Biological Research (Freeman, New York).
- 38.Horiuchi, J., Jiang, W., Zhou, H., Wu, P. & Yin, J. C. (2003) J. Biol. Chem. 279, 12117-12125. [DOI] [PubMed] [Google Scholar]
- 39.Feany, M. B. & Bender, W. W. (2000) Nature 404, 394-398. [DOI] [PubMed] [Google Scholar]
- 40.Gerber, H. P., Seipel, K., Georgiev, O., Hofferer, M., Hug, M., Rusconi, S. & Schaffner, W. (1994) Science 263, 808-811. [DOI] [PubMed] [Google Scholar]
- 41.Mayr, B. & Montminy, M. (2001) Nat. Rev. Mol. Cell. Biol. 2, 599-609. [DOI] [PubMed] [Google Scholar]
- 42.Hansson, O., Nylandsted, J., Castilho, R. F., Leist, M., Jaattela, M. & Brundin, P. (2003) Brain Res. 970, 47-57. [DOI] [PubMed] [Google Scholar]
- 43.Hay, D. G., Sathasivam, K., Tobaben, S., Stahl, B., Marber, M., Mestril, R., Mahal, A., Smith, D. L., Woodman, B. & Bates, G. P. (2004) Hum. Mol. Genet. 13, 1389-1405. [DOI] [PubMed] [Google Scholar]
- 44.Helmlinger, D., Abou-Sleymane, G., Yvert, G., Rousseau, S., Weber, C., Trottier, Y., Mandel, J.-L. & Devys, D. (2004) J. Neurosci. 24, 1881-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang, H., Nucifora, F. C., Jr., Ross, C. A. & DeFranco, D. B. (2003) Hum. Mol. Genet. 12, 1-12. [DOI] [PubMed] [Google Scholar]
- 46.Tanaka, K., Nogawa, S., Nagata, E., Ito, D., Suzuki, S., Dembo, T., Kosakai, A. & Fukuuchi, Y. (2000) Exp. Neurol. 161, 462-471. [DOI] [PubMed] [Google Scholar]
- 47.Irving, E. A., Barone, F. C., Reith, A. D., Hadingham, S. J. & Parsons, A. A. (2000) Brain Res. Mol. Brain Res. 77, 65-75. [DOI] [PubMed] [Google Scholar]
- 48.Crossthwaite, A. J., Hasan, S. & Williams, R. J. (2002) J. Neurochem. 80, 24-35. [DOI] [PubMed] [Google Scholar]
- 49.Hughes, J. P., Staton, P. C., Wilkinson, M. G., Strijbos, P. J., Skaper, S. D., Arthur, J. S. & Reith, A. D. (2003) J. Neurochem. 86, 25-32. [DOI] [PubMed] [Google Scholar]
- 50.Chen, H. K., Fernandez-Funez, P., Acevedo, S. F., Lam, Y. C., Kaytor, M. D., Fernandez, M. H., Aitken, A., Skoulakis, E. M., Orr, H. T., Botas, J., et al. (2003) Cell 113, 457-468. [DOI] [PubMed] [Google Scholar]
- 51.Luthi-Carter, R., Strand, A. D., Hanson, S. A., Kooperberg, C., Schilling, G., La Spada, A. R., Merry, D. E., Young, A. B., Ross, C. A., Borchelt, D. R., et al. (2002) Hum Mol Genet 11, 1927-1937. [DOI] [PubMed] [Google Scholar]
- 52.Chan, E. Y. W., Luthi-Carter, R., Strand, A., Solano, S. M., Hanson, S. A., DeJohn, M. M., Kooperberg, C., Chase, K. O., DiFiglia, M., Young, A. B., et al. (2002) Hum. Mol. Genet. 11, 1939-1951. [DOI] [PubMed] [Google Scholar]
- 53.Sakahira, H., Breuer, P., Hayer-Hartl, M. K. & Hartl, F. U. (2002) Proc. Natl. Acad. Sci. USA 99 Suppl. 4, 16412-16418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muchowski, P. J., Schaffar, G., Sittler, A., Wanker, E. E., Hayer-Hartl, M. K. & Hartl, F. U. (2000) Proc. Natl. Acad. Sci. USA 97, 7841-7846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chan, H. Y., Warrick, J. M., Gray-Board, G. L., Paulson, H. L. & Bonini, N. M. (2000) Hum. Mol. Genet. 9, 2811-2820. [DOI] [PubMed] [Google Scholar]
- 56.Krobitsch, S. & Lindquist, S. (2000) Proc. Natl. Acad. Sci. USA 97, 1589-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wacker, J. L., Zareie, M. H., Fong, H., Sarikaya, M. & Muchowski, P. J. (2004) Nat. Struct. Mol Biol 11, 1215-1222. [DOI] [PubMed] [Google Scholar]
- 58.Freiman, R. N. & Tjian, R. (2002) Science 296, 2149-2150. [DOI] [PubMed] [Google Scholar]
- 59.Helmlinger, D., Bonnet, J., Mandel, J.-L., Trottier, Y. & Devys, D. (2004) J. Biol. Chem. 279, 55969-55977. [DOI] [PubMed] [Google Scholar]
- 60.Schaffar, G., Breuer, P., Boteva, R., Behrends, C., Tzvetkov, N., Strippel, N., Sakahira, H., Siegers, K., Hayer-Hartl, M. & Hartl, F. U. (2004) Mol. Cell 15, 95-105. [DOI] [PubMed] [Google Scholar]
- 61.Agrawal, N., Pallos, J., Slepko, N., Apostol, B. L., Bodai, L., Chang, L.-W., Chiang, A.-S., Thompson, L. M. & Marsh, J. L. (2005) Proc. Natl. Acad. Sci. USA 102, 3777-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yin, J. C., Del Vecchio, M., Zhou, H. & Tully, T. (1995) Cell 81, 107-115. [DOI] [PubMed] [Google Scholar]
- 63.Hendricks, J. C., Williams, J. A., Panckeri, K., Kirk, D., Tello, M., Yin, J. C. & Sehgal, A. (2001) Nat. Neurosci. 4, 1108-1115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.