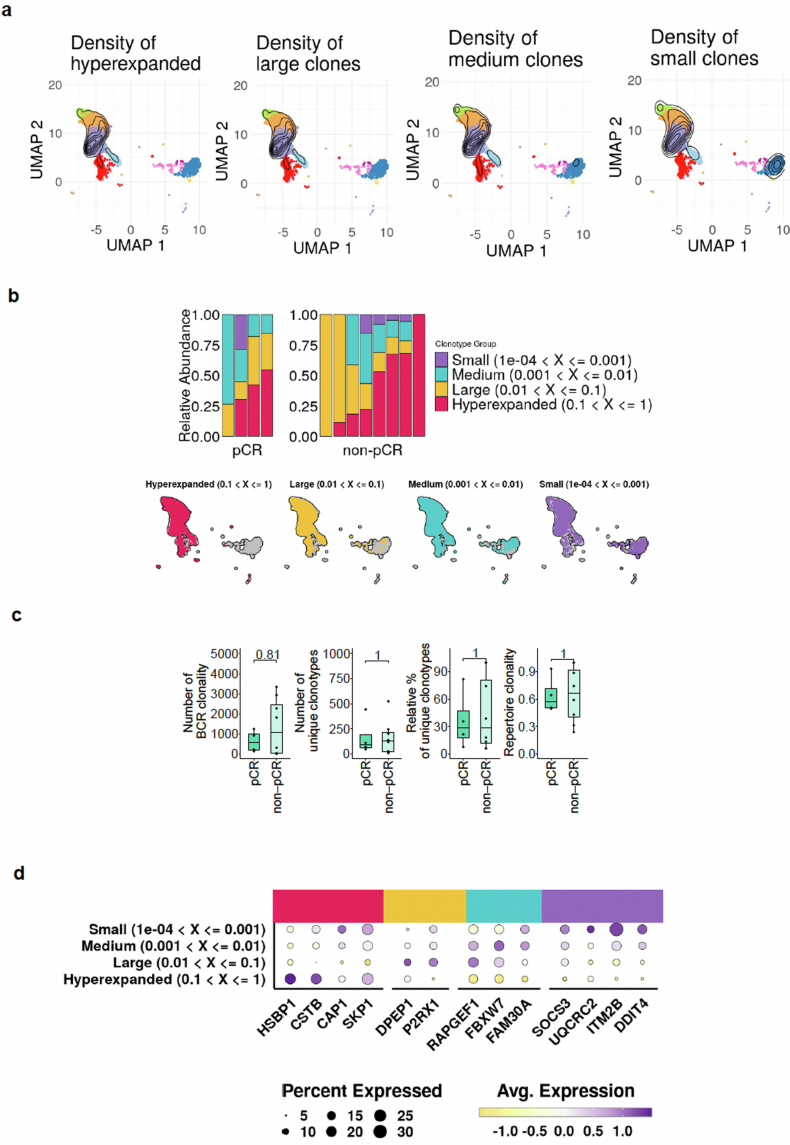
Extended Data Fig. 9. Clonal analysis of B cells in T-VEC-treated basal cell carcinomas.
a, Paired single-cell RNA sequencing (scRNA-seq) and single-cell B cell receptor sequencing (scBCR-seq) demonstrated by uniform manifold approximation and projection (UMAP) showing the density of hyper-expanded, large, medium, and small B and plasma cell clones. b, Bar plots indicating the relative abundance of B and plasma cells by clonality in each paired single-cell RNA sequencing (scRNA-seq) and single-cell B cell receptor sequencing (scBCR-seq) dataset, split by pathological response. UMAPs indicate the distribution of cells by clonality. c, Box plots illustrate the absolute number of clonotypes, the number of unique clonotypes, relative percent of unique clonotypes, and repertoire clonality (calculated as 1 minus normalized entropy [109]) of B and plasma cell clones according to patients with a pathological complete response (pCR) and pathological non-complete response (non-pCR) (n = 12). Data are presented as mean values +/- SEM, p-values are included in individual plots (two-sided Wilcoxon rank-sum test; statistical significance determined with p < 0.05). Each box extends from the 25th (Q1, lower bound of box) to the 75th (Q3, upper bound of box) percentile, the horizontal line in the center of the box represents the median value (Q2). The whiskers extend to the 5th (min, lower bound) and the 95th percentile (max, upper bound). Dots outside the whiskers indicate outliers. d, Bubble plot demonstrating the average expression of differentially expressed (DE) genes in B and plasma cell clones.