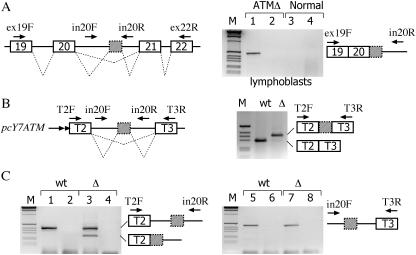
Figure 5.
Analysis of ATM pre-mRNA splicing precursors in lymphoblasts and hybrid minigenes. (A) The left panel shows a schematic representation of the human ATM gene and the location of the primers used for RT–PCR analysis. Exons are indicated as boxes, introns as line and dotted lines represent the possible splicing products. The cryptic ATM exon is shown as a grey box with dotted outline. Total RNA, extracted from ATM patient (ATMΔ) and normal lymphoblasts, was treated with DNAse RNAse-free, reverse transcribed and amplified with the indicated primers. Lanes 1 and 3 correspond to amplification with ex19F and in20R, lanes 2 and 4 to amplification with in20F and ex22R. The identity of the amplified precursor product detected in the affected lymphoblasts which contains exon 19, exon 20, the ATMΔ cryptic exon and the downstream intron 20 sequences is schematically represented on the right on the splicing assay along the primers utilized. Control experiments without reverse transcriptase did not reveal any amplified band (data not shown). M is the molecular weight marker 1 kb. (B) Schematic representation of the short α-tropomyosin hybrid minigene (pcY7ATM) used in transient transfection assay. Tropomyosin exons 2 (T2) and 3 (T3) and the ATM cryptic exon (grey) are boxed. The length of the intronic sequences (shown as lines and not in scale) flanking the ATM cryptic exon is of 99 and 108 bases, respectively. Dotted lines represent the possible splicing products. pcY7ATM wt and Δ minigenes were transiently transfected in Hep3B cells and the pattern of splicing analyzed with the indicated primers. Amplification experiments of mature mRNA with T2F and T3R is shown on the right. As expected the GTAA deletion activates the cryptic exon also in the short context of the α-tropomyosin gene. (C) Result of amplification experiments of pcY7ATM wt and Δ with T2F-in20R and in20F-T3R primers on the left and right panel, respectively. The pre-mRNA forms are schematically represented along with the primers utilized. Even lanes correspond to control amplification of RNA samples without reverse transcriptase. All amplified bands were verified by sequencing.