Abstract
The generation of microarray probes with specificity below the species level is an ongoing challenge, not least because the high-throughput detection of microorganisms would be an efficient means of identifying environmentally relevant microbes. Here, we describe how suppression subtractive hybridization (SSH) can be applied to the production of microarray probes that are useful for microbial differentiation at the subspecies level. SSH was used to initially isolate unique genomic sequences of nine Salmonella strains, and these were validated in quadruplicate by microarray analysis. The results obtained indicate that a large group of genes subtracted by SSH could serve together, as one probe, for detecting a microbial subspecies. Similarly, the whole microbial genome (not subjected to SSH) can be used as a species-specific probe. The detailed methods described herein could be used and adapted for the estimation of any cultivable bacteria from different environments.
INTRODUCTION
Microarrays are powerful tools for the parallel, high-throughput detection and quantification of nucleic acids (1). Currently, DNA microarrays, in particular, are broadly applied across most sectors of the life sciences, including environmental microbiology and microbial ecology and human, veterinary, food and plant diagnostics (2,3). While there is abundant opportunity to use DNA microarray technology to identify microorganisms, several practical limitations have slowed the implementation of this methodology (4). These include low sensitivity and poor resolution at the species and subspecies level. For the potential of DNA microarrays to be realized, these long-term methodological obstacles must be overcome. Because species-level resolution does not necessarily provide sufficient information for diagnosis, there is a need for subspecies-level genetic resolution markers in both clinical and environmental microbiology (1). In this study, sets of genes produced by suppression subtractive hybridization (SSH) were tested as subspecies-specific microarray probes for discriminating several strains that are closely related phylogenetically.
SSH is a widely used method for separating DNA sequences that distinguish two closely related genomic DNA (gDNA) libraries (5–8). A key feature of the method is simultaneous normalization and subtraction steps. The normalization step equalizes the abundance of DNA fragments within the target population, and the subtraction step excludes sequences that are common to the two populations being compared (7). Specific amplification of genes with SSH has allowed identification of minute genomic differences between closely related microbial strains (9–13) and enabled the profiling of genetic diversity in an environmental metagenome (14). Although there have been several studies applying tester-specific genes as microarray probes (15–17), they all used a single clone as an individual probe because their microarray was constructed for analysis of genome-wide differential gene expression. Recently, using nylon membrane-based macroarray hybridization, Li et al. (18) demonstrated that gene fragments produced by SSH could be used as a species-specific probe for the diagnosis of five species of the genus Dendrobrium. While that study provided valuable insights into gDNA subtraction between different species, we suggest that their approach may actually be subspecies-specific, given that the gDNA probes were absolutely species-specific in the reverse sample genome probing (RSGP) hybridization (19). In addition, while those authors reported that only 12.16% of SSH clones were species-specific, it is generally recognized that all fragments from SSH are absolutely tester-specific (14). In this study, therefore, SSH-amplified genes that are specific to a microbial strain were not separated into individual fragments by cloning. Rather, the sum of amplified genes was applied together as one microarray probe, which was used to specifically detect the individual strain. The diagnostic specificity of these microarray probes was evaluated by typing nine Salmonella type strains that are phylogenetically closely related and by comparing them to gDNA probes. For more accurate and precise analysis, miniaturized microarrays with guaranteed high reproducibility (20) were constructed by depositing DNA probes onto non-porous substrates with printing robots. The underlying rationale of SSH-microarray hybridization is discussed from a phylogenomic perspective.
MATERIALS AND METHODS
Bacterial strains and preparation of gDNA
Nine Salmonella type strains plus an Escherichia coli type strain were used in this study, as detailed in Table 1. They were obtained from the Korean Collection for Type Cultures (KCTC), the German Collection of Microorganisms and Cell Cultures (DSMZ) and the American Type Culture Collection (ATCC). All strains were maintained and grown under conditions suggested by the collection from which they were sourced. Cells at the exponential growth phase were quickly harvested and frozen at −80°C for the extraction of DNA. The gDNAs were isolated using a bead-beating method, as previously described (21). All DNA samples were treated with RNase A (Sigma, St Louis, MO) and were analyzed on agarose gels stained with ethidium bromide prior to SSH, microarray fabrication and hybridization. Concentrations of the obtained DNAs were determined in triplicate using a spectrophotometer (Nanodrop Technologies, Rockland, DE).
Table 1.
Microorganisms whose genomes were used as microarray probes
| Location in microarraya | Target microorganism used as a SSH tester | SSH driver | ||
|---|---|---|---|---|
| Genus and species name | Subspecies name | Culture collection number | ||
| A | S.choleraesuis | indica | DSM 14848 | S.choleraesuis subsp. choleraesuis |
| B | S.choleraesuis | choleraesuis | DSM 14846 | S.typhimurium |
| C | S.bongori | – | DSM 13772 | S.choleraesuis subsp. choleraesuis |
| D | S.choleraesuis | arizonae | DSM 9386 | S.choleraesuis subsp. choleraesuis |
| E | S.choleraesuis | hountenae | DSM 9221 | S.choleraesuis subsp. choleraesuis |
| F | S.choleraesuis | salamae | DSM 9220 | S.choleraesuis subsp. choleraesuis |
| G | S.typhimurium | – | ATCC 13311 | S.choleraesuis subsp. choleraesuis |
| H | S.enteritidis | – | ATCC 13076 | S.choleraesuis subsp. choleraesuis |
| I | S.choleraesuis | diarizonae | DSM 14847 | S.choleraesuis subsp. choleraesuis |
| b | E.coli | – | KCTC 2441 | S.choleraesuis subsp. choleraesuis |
aAll gene probes were printed in quadruplicate. Microbial gDNAs were printed in the first row, and SSH probes in the second row (Figure 3).
bThe gDNA and SSH probes for E.coli were not printed together in the same section as those for Salmonella, in the interests of clarity.
Suppressive subtractive hybridization
SSH was used to isolate DNA fragments present in the target microbial organisms but absent from the reference strains. The procedure was performed using the PCR-Select Bacterial Genome Subtraction Kit (Clontech), with minor modifications. Salmonella choleraesuis subsp. choleraesuis (Type species) was assigned as the SSH driver while the other eight strains were assigned as SSH testers. For SSH of S.choleraesuis subsp. choleraesuis, Salmonella typhimurium, which has the most similar 16S rRNA sequence, was assigned as a driver. Tester and driver samples were digested separately with RsaI. Adaptor ligations using the Clontech kit were performed as recommended in the user manual. Ligation efficiency was analyzed also as described in the manual with the bacterial 1088r primer (22). PCR amplification of tester-specific fragments was performed using primers directed to tester ligated adaptor sequences. The AccuPower® PCR PreMix (Bioneer, Korea) and a Peltier Thermal Cycler (DNA Engine DYAD™, MJ Research) were used for both primary and secondary PCR amplifications. Both PCR amplifications were performed using the primers and recommended primer and template concentrations from the Clontech kit, adjusted to a total volume of 50 μl. The primary PCR cycling conditions were as follows: one incubation of 94°C for 5 min followed by 30 cycles of 94°C for 10 s, 66°C for 30 s and 72°C for 1.5 min. Secondary PCR cycling involved a similar program, but used an annealing temperature of 68°C. Five replicates of each secondary PCR product (250 μl) were purified using the AccuPrep® PCR Purification Kit (Bioneer, Korea).
Microarray construction, labeling and hybridization
Microarray construction, post-treatment, labeling and hybridization were performed as described previously (23) with minor modifications. The gDNAs and genes specifically amplified by SSH were diluted to a final concentration of 400 ng/μl in 0.1× TE. Five microliters aliquots of each probe were transferred to a 384-well microplate and mixed with 5 μl of 2× microarray spotting solution (ArrayIt™, Telechem International, Inc., Sunnyvale, CA) for printing. Each probe set was printed in quadruplicate. The exact location of each gDNA on the glass slide is listed in Table 1. After post-treatments including cross-linking, blocking, heat-denaturing (immersing in dH2O at 95°C for 2 min), rinsing in 95% ethanol and air-drying, microarrays were stored dry in a clean slide box at room temperature. DNA was labeled with the BioPrime DNA Labeling System and 2.5 mM Cy5 dCTP (Amersham Pharmacia Biotech, Piscataway, NJ) by incubating at 37°C for 3 h. The labeled target DNA was purified using a QIAquick PCR purification column (Qiagen, Valencia, CA), concentrated in a Speedvac for 1 h, and resuspended in 4.35 μl of dH2O for hybridization. All microarray hybridizations were performed in triplicate (a total of 12 replicates per DNA probe), unless otherwise noted, to facilitate statistical analyses. After hybridization, each microarray slide was taken out and its cover slip was immediately removed under wash solution 1 (1× SSC and 0.2% SDS). Slides were washed using wash solution 1, wash solution 2 (0.1× SSC and 0.2% SDS), and wash solution 3 (0.1× SSC) for 5 min each, at ambient temperature prior to drying. The slide was dried by centrifugation, as described above.
Microarray scanning and data analysis
A GenePix® 4000B microarray scanner set (Axon instruments, Union City, CA) was used for scanning the microarrays at a resolution of 10 μm. For consistent scanning of all hybridized slides, the laser power and photomultiplier tube (PMT) gain were adjusted to 100%. Scanned image displays were analyzed by quantifying the pixel density (intensity) of each hybridization spot using the GenePix® software program (version 4.0; Axon instruments). Hybridization images presented here are representative images, which were automatically contrast-adjusted by the software. A grid of individual circles defining the location of each DNA spot on the array was superimposed onto the image to indicate each quantified fluorescent spot. Mean signal intensity was automatically determined for each spot. The local background signal was also automatically subtracted from the hybridization signal of each individual spot. The signal-to-noise ratio (SNR) for each spot was calculated based on following formula (24): SNR = (signal intensity − background)/SD of background, in which the ‘background’ measurement refers to the local spot background intensity and the ‘SD of background’ was calculated across all pixels, as measured by the GenePix® software. Statistical analysis was performed using Excel 2003 (Microsoft) and Sigmaplot 8.0 (Jandel Scientific, San Rafael, CA). The SNRs from 12 replicate data sets were then averaged to obtain the SNR value of each individual probe. Relative SNRs were obtained by dividing the average SNR value by the mean value of the hybridized template SNR in the same microarray slide.
Phylogenetic analysis of the genus Salmonella
16S rDNA isolated from each Salmonella strain was amplified by PCR using two universal primers as described by Yoon et al. (25) and sequenced as described by Yoon et al. (26). Multiple alignments were performed using the Clustal X program (27). The evolutionary distances were calculated using the Jukes & Cantor method. The phylogenic tree was constructed using a neighbor-joining method (28) in the MEGA 2 program (29). To compare similarities, a 16S rRNA homology matrix was constructed using PairProWin.exe (http://microarray.kaist.ac.kr) software, which is based on the Clustal W program.
RESULTS
Phylogenetic analysis on the genus Salmonella
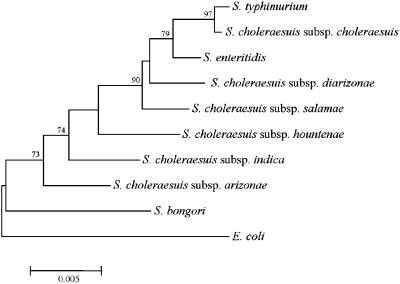
Currently, the valid names of species and subspecies in the genus Salmonella are as follows: S.choleraesuis (subsp. choleraesuis, salamae, arizonae, diarizonae, hountenae, and indica), S.enteritidis, S.typhimurium, Salmonella typhi and Salmonella bongori (30). Although Salmonella enteritidis, S.typhimurium and S.typhi are almost genetically identical to S.choleraesuis subsp. choleraesuis (31–33), these species continue to appear in the approved lists and retain their unique nomenclatural because they are now very important human pathogens. In this study, the type strains of S.enteritidis, S.typhimurium, S.bongori and six subspecies (serovars) of S.choleraesuis were selected to determine whether SSH-generated microarray probes are specific at a subspecies level. To confirm the identity of these strains and to preliminarily investigate phylogenetic relationships within the genus Salmonella, 16S rDNA was sequenced for each strain, subjected to BLAST searching (34) and inter-strain comparisons were conducted. This 16S rDNA gene-based phylogeny revealed that the Salmonella type strains used in this study are very closely related with all species and subspecies, except S.bongori, sharing >98% 16S rDNA sequence similarity. S.bongori had 97.2–98.0% sequence identity with the other Salmonella type strains while E.coli, which was used as an experimental control and an out-group in the phylogenetic tree, had 96.7–97.9% similarity with Salmonella. The 16S rDNA distances between the Salmonella strains used in this study are plotted in Figure 1.
Figure 1.
Neighbor-joining tree based on 16S rDNA sequences showing the phylogenetic position of each type strain used in this study in the genus Salmonella. Bootstrap values (1000 replications) are shown as percentages at each node only if they are 50% or greater. Scale bar represents 0.005 substitutions per nucleotide position. E.coli was used as an out-group.
SSH of gDNA from Salmonella species and subspecies
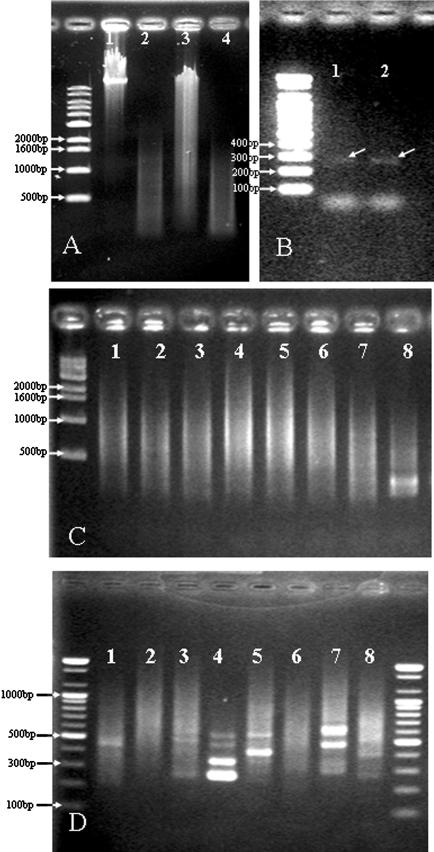
SSH was carried out between the type strains of the nine Salmonella species and subspecies to determine the usefulness of this technique for isolating DNA unique to a Salmonella subspecies. S.choleraesuis subsp. choleraesuis (type species of the genus) was assigned as the SSH driver while the other eight strains were assigned as SSH testers. Based on 16S rRNA homology comparison, the nearest phylogenetic neighbor, S.typhimurium, was assigned as the driver for S.choleraesuis subsp. choleraesuis (Figure 1). Most tester and driver samples were between 100 and 1500 bp in size when digested with RsaI (Figure 2A). Analysis of ligation efficiency, which was conducted by PCR amplification with the bacterial 1088r primer, produced a dim band of ∼280 bp in size. This corresponded to the fragment size predicted on the basis of the priming and RsaI-cutting sites in the Salmonella 16S rRNA sequence (Figure 2B). After twice hybridization between testers and drivers, primary PCR and secondary nested PCR of tester-specific fragments were executed. Although the apparent intensity and size of ethidium bromide-stained primary products were similar for all tester species/subspecies, the size of secondary nested PCR products varied significantly between testers (Figure 2C and D). Concentrations of the purified secondary PCR products (50 μl) were in the range 62–119 ng/μl. These products were further concentrated and used as probes for SSH microarrays.
Figure 2.
The SSH procedure. (A) Examples of extracted gDNA (1: S.choleraesuis subsp. choleraesuis, 3: S.typhimurium) and RsaI-digested DNA (2,4). (B) Examples of the adaptor ligation efficiency test. PCR amplification was executed with an adaptor primer and the bacterial 1088r primer. (C) Examples of primary PCR results. (D) Examples of secondary nested PCR results. Numbers of testers (1–8) shown in C and D are exactly the same as the microarray location numbers.
Specificity of SSH microarrays
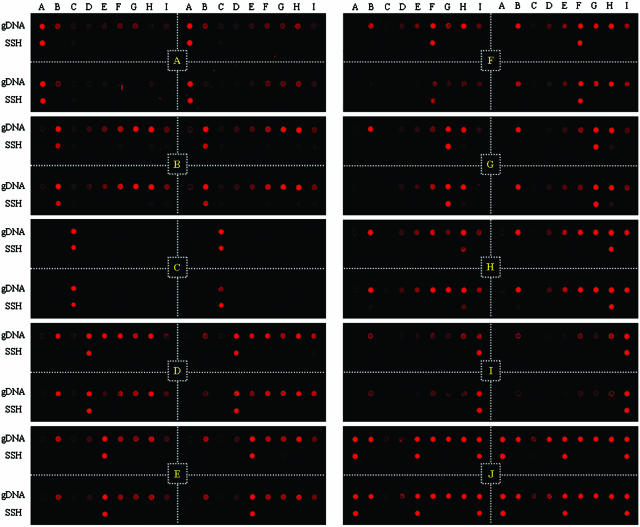
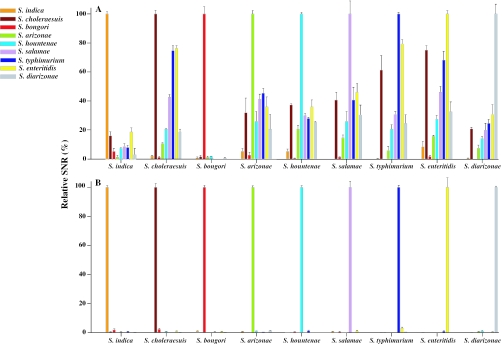
To obtain proof of principle, the specificity of SSH microarrays was evaluated by hybridizing them with the gDNA of each Salmonella type strain. As predicted, strong signals were only obtained for the probes corresponding to the labeled target (lower lines in Figure 3). Little or no cross-hybridization (0–4%) of SNRs was observed for non-target species and subspecies, indicating that, at least under the conditions applied here, strain-specific hybridization is achieved with SSH microarrays. In order to compare the specificity of SSH probes to intact genome probes, gDNA extracted from each genus Salmonella strain was also spotted on the same slide (upper lines in Figure 3). In the Salmonella gDNA hybridization, cross-hybridizations ranging from 1 to 92% of (SNRs) were observed between the target subspecies and its phylogenetic neighbor subspecies. Different species such as S.bongori and E.coli were clearly distinguished by genomic hybridization whereas almost identical species, such as S.typhimurium and S.enteritidis, could not be differentiated from S.choleraesuis subsp. choleraesuis (31). This indicated that strain or subspecies differentiation could not be achieved in gDNA-based hybridization, in contrast to the SSH probes, all of which were perfectly specific for their corresponding targets. Regarding cross-hybridization of gDNA probes, there was a clear trend for higher cross-hybridization SNR values among subspecies with close 16S rDNA homology (Figures 1 and 4). This strongly suggests that genomic hybridization on a microarray slide is very useful for estimating bacterial similarity, below the species level, for closely related strains.
Figure 3.
Fluorescence images showing hybridization specificity of gDNA and SSH probes. All gene probes were printed in quadruplicate (divided with white dotted lines). Microbial gDNAs were printed in the first row (gDNA), and SSH probes in the second row (SSH). Location of probes in the microarray is shown in Table 1. Each microarray was hybridized with 500 ng of Salmonella gDNA (see yellow indications on each image and Table 1), except the 3 mix. The microarray indicated as J (yellow indication) was hybridized with 500 ng of equally mixed gDNAs of S.choleraesuis subsp. indica (A), S.choleraesuis subsp. hountenae (E) and S.choleraesuis subsp. diarizonae (I).
Figure 4.
Histogram plots showing relative SNRs of the hybridization specificity images (Figure 3) of gDNA (A) and SSH probes (B). The probe sources of gDNA and SSH tester were indicated in the left of the figure and hybridized templates were shown below the grouped bars.
Since real environments are generally composed of a variety of different microorganisms, evaluation of SSH probes with mixed genomes may enhance the applicability of SSH microarrays. To determine whether other non-target DNA interferes with the specificity of SSH microarray-based hybridization, three different gDNAs (500 ng per species) from S.choleraesuis subsp. indica, hountenae and diarizonae were mixed, labeled, and hybridized to the SSH microarray (J in Figure 3). Specific hybridization signals were observed for the corresponding probes, with these exhibiting similar SNRs of 286.3, 292.8 and 309.4, respectively. On the contrary, the non-target probes had SNRs below the background level. These results suggest that SSH probes could also be specific in the presence of other genomes and strongly validate the hypothesis that the subtracted genome only contains tester-specific genes.
DISCUSSION
Salmonella is one of the major bacterial agents that cause foodborne infections in humans worldwide (35). Members of the genus Salmonella have been divided into >2300 serovars (32) and their classification has provoked many arguments between microbiologists (30). The extraordinarily close inter-genomic relationships that exist between Salmonella species present a major problem for prohibiting their phylogeny from being unshakable (36). Since the major epidemiological markers of Salmonella strains are subspecies and serovars, which are determined by chromosomal genes and are not affected by extrachromosomal elements (37), high-throughput detection of Salmonella below the species level is crucial for studying human infections in detail. Existing methods for the molecular typing of Salmonella include plasmid typing, pulsed field gel electrophoresis, ribotyping and random-amplified polymorphic DNA analyses (38). However, the ever-increasing availability of genome sequences for these microorganisms implies that genotypic methods that are amenable to automation will be developed in time. In this regard it is plausible that DNA microarrays will be developed with specific probes for thousands of different microbial species or strains, enabling fast and reliable identification of microorganisms (39). Most previous Salmonella microarrays have been manufactured for the purpose of studying genetic relationships between Salmonella and/or other close relatives (40). Although a few 16S rRNA gene-based microarrays have been developed for high-throughput diagnosis of bacterial pathogens including Salmonella (41,42), so far these have exhibited poor resolution at the species level as well as problems in detection sensitivity (43,44).
In this study, we differentiated seven subspecies of Salmonella using SSH-generated microarray probes. Their ability to distinguish S.enteritidis and S.typhimurium from the almost genetically identical organism, S.choleraesuis subsp. choleraesuis (and vice versa), indicated that SSH probes are strain-specific. Importantly, this specificity was also maintained in a mixed genome hybridization test. These results indicate that the generation and application of microarray probes with SSH may be useful for monitoring various microorganisms in real environments. Although Li et al. (18) reported that only 12.16% of SSH clones were species-specific, all the SSH fragments in this study were found to be absolutely tester-specific since no cross-hybridization was observed, even between almost identical strains. The high degree of specificity of SSH probes was not surprising, given that gDNA probes were also absolutely species-specific—an observation consistent with previous findings for RSGP hybridization (19).
Subtracting the tester genome with a driver genome is not an ideal approach for the construction of strain-specific probes, since it is possible that the remaining tester strain gDNA could still contain the genes similar to those of a third unknown organism. The potential paucity of SSH microarrays could be addressed by selecting the most closely related strain as a SSH driver for each test strain and using a nested probe approach such that probes of different taxonomic levels, such as gDNA, are combined to increase the confidence of the assay.
The use of microbial diagnostic microarrays is expected to accelerate our understanding of previously unrecognized complex microbial processes (2). However, high resolution microarray analysis techniques must also be developed on a high-throughput basis. This will provide a significant advance over current techniques since for many organisms, even species-level differentiation is difficult to achieve (1). The specificity of SSH probes observed in this study could clearly solve the existing methodological problems associated with microarrays. The ability to conduct such high resolution analyses has important implications across diverse areas including the monitoring of inoculated strains in bioreactors or the lactic acid bacteria used as a probiotics in gastrointestinal microflora, as well as the diagnosis of closely related microorganisms such as Salmonella.
Acknowledgments
This work was supported by grant BDM0200413, grant NNM0100411 and the NRL research programme (grant M10104000294-01J000012800) from the Korean Ministry of Science and Technology (MOST). Funding to pay the Open Access publication charges for this article was provided by KRIBB Research Initiative Program.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bodrossy L., Sessitsch A. Oligonucleotide microarrays in microbial diagnostics. Curr. Opin. Microbiol. 2004;7:245–254. doi: 10.1016/j.mib.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Zhou J. Microarrays for bacterial detection and microbial community analysis. Curr. Opin. Microbiol. 2003;6:288–294. doi: 10.1016/s1369-5274(03)00052-3. [DOI] [PubMed] [Google Scholar]
- 3.Zhou J., Thompson D.K. Challenges in applying microarrays to environmental studies. Curr. Opin. Biotechnol. 2002;13:204–207. doi: 10.1016/s0958-1669(02)00319-1. [DOI] [PubMed] [Google Scholar]
- 4.Cook K.L., Sayler G.S. Environmental application of array technology: promise, problems and practicalities. Curr. Opin. Biotechnol. 2003;14:311–318. doi: 10.1016/s0958-1669(03)00057-0. [DOI] [PubMed] [Google Scholar]
- 5.Rebrikov D.V., Desai S.M., Siebert P.D., Lukyanov S.A. Suppression subtractive hybridization. Methods Mol. Biol. 2004;258:107–134. doi: 10.1385/1-59259-751-3:107. [DOI] [PubMed] [Google Scholar]
- 6.Lukyanov S.A., Gurskaya N.G., Lukyanov K.A., Tarabykin V.S., Sverdlov E.D. Highly efficient subtractive hybridization of cDNA. J. Bioorg. Chem. 1994;20:386–388. [Google Scholar]
- 7.Gurskaya N.G., Diatchenko L., Chenchik A., Siebert P.D., Khaspekov G.L., Lukyanov K.A., Vagner L.L., Ermolaeva O.D., Lukyanov S.A., Sverdlov E.D. Equalizing cDNA subtraction based on selective suppression of polymerase chain reaction: cloning of Jurkat cell transcripts induced by phytohemaglutinin and phorbol 12-myristate 13-acetate. Anal. Biochem. 1996;240:90–97. doi: 10.1006/abio.1996.0334. [DOI] [PubMed] [Google Scholar]
- 8.Diatchenko L., Lau Y.F., Campbell A.P., Chenchik A., Moqadam F., Huang B., Lukyanov S., Lukyanov K., Gurskaya N., Sverdlov E.D., Siebert P.D. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl Acad. Sci. USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harms C., Kase M., Hildebrandt A. Characterization of minute differences between genomes of strains of Penicillium nalgiovense using subtractive suppression hybridization without cloning. Lett. Appl. Microbiol. 2002;35:113–116. doi: 10.1046/j.1472-765x.2002.01145.x. [DOI] [PubMed] [Google Scholar]
- 10.Stocki S.L., Babiuk L.A., Rawlyk N.A., Potter A.A., Allan B.J. Identification of genomic differences between Escherichia coli strains pathogenic for poultry and E.coli K-12 MG1655 using suppression subtractive hybridization analysis. Microb. Pathog. 2002;33:289–298. doi: 10.1006/mpat.2002.0536. [DOI] [PubMed] [Google Scholar]
- 11.Harakava R., Gabriel D.W. Genetic differences between two strains of Xylella fastidiosa revealed by suppression subtractive hybridization. Appl. Environ. Microbiol. 2003;69:1315–1319. doi: 10.1128/AEM.69.2.1315-1319.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai Z., Mao X., Magnuson J.K., Lasure L.L. Identification of genes associated with morphology in Aspergillus niger by using suppression subtractive hybridization. Appl. Environ. Microbiol. 2004;70:2474–2485. doi: 10.1128/AEM.70.4.2474-2485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsieh W.J., Pan M.J. Identification Leptospira santarosai serovar shermani specific sequences by suppression subtractive hybridization. FEMS Microbiol. Lett. 2004;235:117–124. doi: 10.1016/j.femsle.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 14.Galbraith E.A., Antonopoulos D.A., White B.A. Suppressive subtractive hybridization as a tool for identifying genetic diversity in an environmental metagenome: the rumen as a model. Environ. Microbiol. 2004;6:928–937. doi: 10.1111/j.1462-2920.2004.00575.x. [DOI] [PubMed] [Google Scholar]
- 15.Rishi A.S., Munir S., Kapur V., Nelson N.D., Goyal A. Identification and analysis of safener-inducible expressed sequence tags in Populus using a cDNA microarray. Planta. 2004;220:296–306. doi: 10.1007/s00425-004-1356-9. [DOI] [PubMed] [Google Scholar]
- 16.Pomati F., Burns B.P., Neilan B.A. Identification of an Na(+)-dependent transporter associated with saxitoxin-producing strains of the cyanobacterium Anabaena circinalis. Appl. Environ. Microbiol. 2004;70:4711–4719. doi: 10.1128/AEM.70.8.4711-4719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munir S., Singh S., Kaur K., Kapur V. Suppression subtractive hybridization coupled with microarray analysis to examine differential expression of genes in virus infected cells. Biol. Proced. Online. 2004;6:94–104. doi: 10.1251/bpo77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li T.X., Wang J., Bai Y., Sun X., Lu Z. A novel method for screening species-specific gDNA probes for species identification. Nucleic Acids Res. 2004;32:e45. doi: 10.1093/nar/gnh041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greene E.A., Voordouw G. Analysis of environmental microbial communities by reverse sample genome probing. J. Microbiol. Methods. 2003;53:211–219. doi: 10.1016/s0167-7012(03)00024-1. [DOI] [PubMed] [Google Scholar]
- 20.Dorris D.R., Ramakrishnan R., Trakas D., Dudzik F., Belval R., Zhao C., Nguyen A., Domanus M., Mazumder A. A highly reproducible, linear, and automated sample preparation method for DNA microarrays. Genome Res. 2002;12:976–984. doi: 10.1101/gr.227402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeates C., Gillings M.R., Davison A.D., Altavilla N., Veal D.A. Methods for microbial DNA extraction from soil for PCR amplification. Biol. Proced. Online. 1998;1:40–47. doi: 10.1251/bpo6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang Y.-H., Kim J.-K., Kim H.-J., Kim W.-Y., Kim Y.-B., Park Y.-H. Selection of a potential probiotic Lactobacillus strain and subsequent in vivo studies. Antonie Van Leeuwenhoek. 2001;80:193–199. doi: 10.1023/a:1012213728917. [DOI] [PubMed] [Google Scholar]
- 23.Rhee S.K., Liu X., Wu L., Chong S.C., Wan X., Zhou J. Detection of genes involved in biodegradation and biotransformation in microbial communities by using 50-mer oligonucleotide microarrays. Appl. Environ. Microbiol. 2004;70:4303–4317. doi: 10.1128/AEM.70.7.4303-4317.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zong Y., Wang Y., Zhang S., Shi Y. How to evaluate a microarray scanner. In: Hardiman G., editor. Microarrays Methods and Applications: Nuts & Bolts. LLC: DNA press; 2003. pp. 97–114. [Google Scholar]
- 25.Yoon J.H., Lee S.T., Park Y.H. Inter- and intraspecific phylogenetic analysis of the genus Nocardioides and related taxa based on 16S rDNA sequences. Int. J. Syst. Bacteriol. 1998;48:187–194. doi: 10.1099/00207713-48-1-187. [DOI] [PubMed] [Google Scholar]
- 26.Yoon J.H., Kim H., Kim I.G., Kang K.H., Park Y.H. Erythrobacter flavus sp. nov., a slight halophile from the East Sea in Korea. Int. J. Syst. Evol. Microbiol. 2003;53:1169–1174. doi: 10.1099/ijs.0.02510-0. [DOI] [PubMed] [Google Scholar]
- 27.Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 29.Kumar S., Tamura K., Jakobsen I.B., Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 30.Ezaki T., Kawamura Y., Yabuuchi E. Recognition of nomenclatural standing of Salmonella typhi (Approved Lists 1980), Salmonella enteritidis (Approved Lists 1980) and Salmonella typhimurium (Approved Lists 1980), and conservation of the specific epithets enteritidis and typhimurium. Request for an opinion. Int. J. Syst. Evol. Microbiol. 2000;50:945–947. doi: 10.1099/00207713-50-2-945. [DOI] [PubMed] [Google Scholar]
- 31.Crosa J.H., Brenner D.J., Ewing W.H., Falkow S. Molecular relationship among the Salmonella. J. Bacteriol. 1973;115:307–315. doi: 10.1128/jb.115.1.307-315.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christensen H., Nordentoft S., Olsen J.E. Phylogenetic relationships of Salmonella based on rRNA sequences. Int. J. Syst. Bacteriol. 1998;48:605–610. doi: 10.1099/00207713-48-2-605. [DOI] [PubMed] [Google Scholar]
- 33.Stoleru G.H., Le Minor L., Lheritier M. Polynucleotide sequence divergence among strains of Salmonella subgenus IV and closely related organisms. Ann. Microbiol. 1976;127:447–486. [PubMed] [Google Scholar]
- 34.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 35.Herikstad H., Motarjemi Y., Tauxe R.V. Salmonella surveillance: a global survey of public health serotyping. Epidemiol Infect. 2002;129:1–8. doi: 10.1017/s0950268802006842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Le Minor L. The Genus Salmonella. 2nd edn. NY: Springer-Verlag; 1991. [Google Scholar]
- 37.Le Minor L. Typing of Salmonella species. Eur. J. Clin. Microbiol. Infect. Dis. 1988;7:214–218. doi: 10.1007/BF01963091. [DOI] [PubMed] [Google Scholar]
- 38.Farber J.M. An introduction to the hows and whys of molecular typing. J. Food Prot. 1996;59:1091–1101. doi: 10.4315/0362-028X-59.10.1091. [DOI] [PubMed] [Google Scholar]
- 39.Kuipers O.P. Genomics for food biotechnology: prospects of the use of high-throughput technologies for the improvement of food microorganisms. Curr. Opin. Biotechnol. 1999;10:511–516. doi: 10.1016/s0958-1669(99)00019-1. [DOI] [PubMed] [Google Scholar]
- 40.Porwollik S., Wong R.M., McClelland M. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc. Natl Acad. Sci. USA. 2002;99:8956–8961. doi: 10.1073/pnas.122153699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Panicker G., Call D.R., Krug M.J., Bej A.K. Detection of pathogenic Vibrio spp. in shellfish by using multiplex PCR and DNA microarrays. Appl. Environ. Microbiol. 2004;70:7436–7444. doi: 10.1128/AEM.70.12.7436-7444.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Call D.R., Borucki M.K., Loge F.J. Detection of bacterial pathogens in environmental samples using DNA microarrays. J. Microbiol. Methods. 2003;53:235–243. doi: 10.1016/s0167-7012(03)00027-7. [DOI] [PubMed] [Google Scholar]
- 43.Dahllof I. Molecular community analysis of microbial diversity. Curr. Opin. Biotechnol. 2002;13:213–217. doi: 10.1016/s0958-1669(02)00314-2. [DOI] [PubMed] [Google Scholar]
- 44.Kakinuma K., Fukushima M., Kawaguchi R. Detection and identification of Escherichia coli, Shigella, and Salmonella by microarrays using the gyrB gene. Biotechnol. Bioeng. 2003;83:721–728. doi: 10.1002/bit.10709. [DOI] [PubMed] [Google Scholar]