Abstract
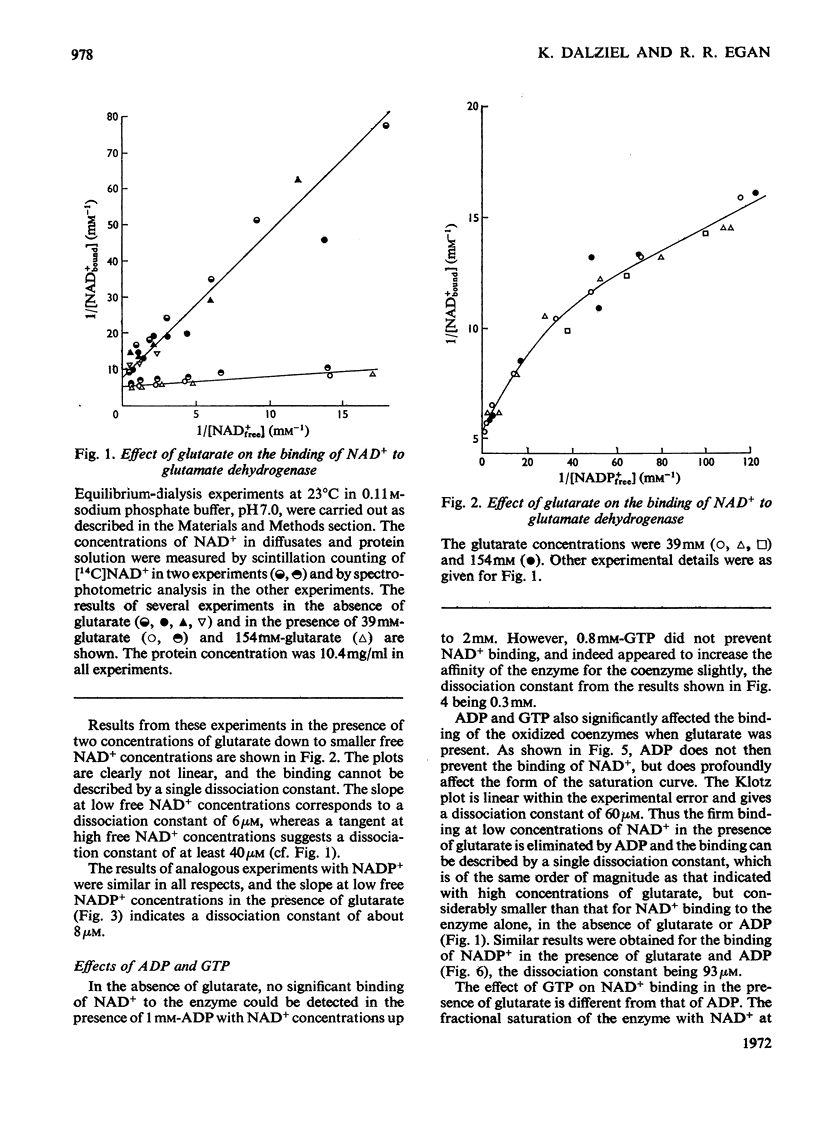
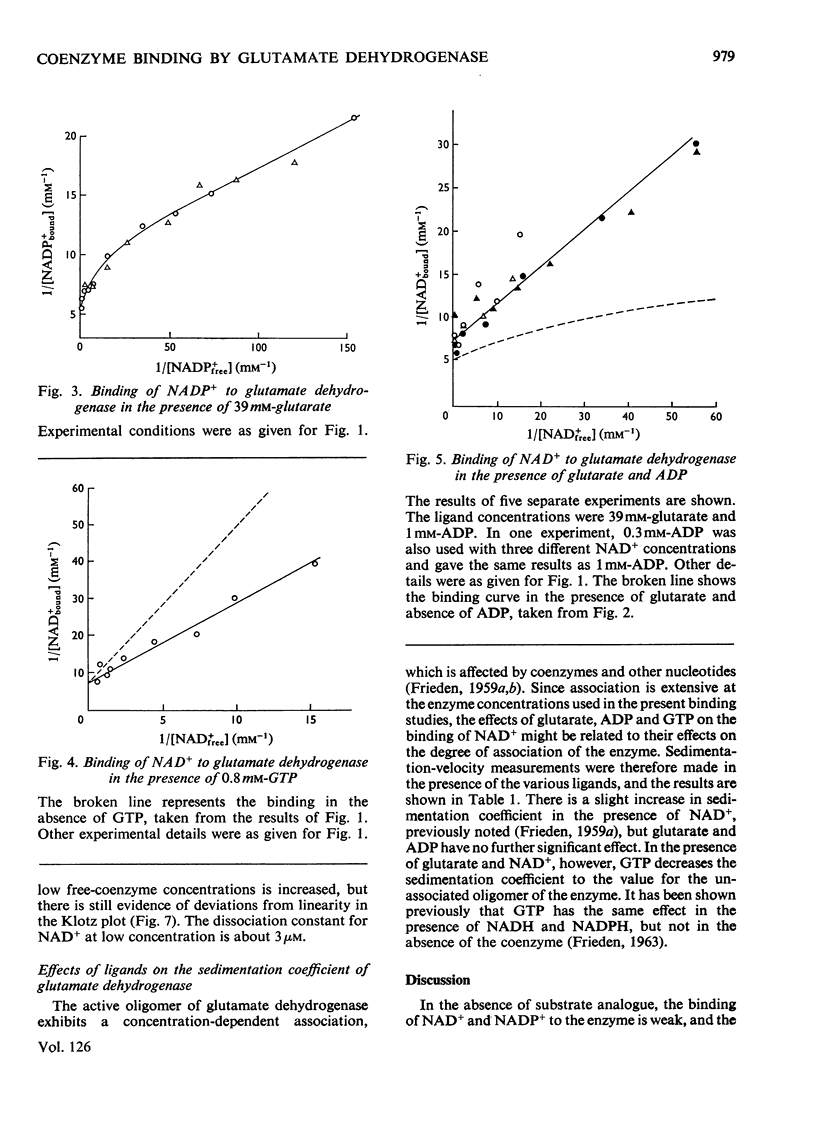
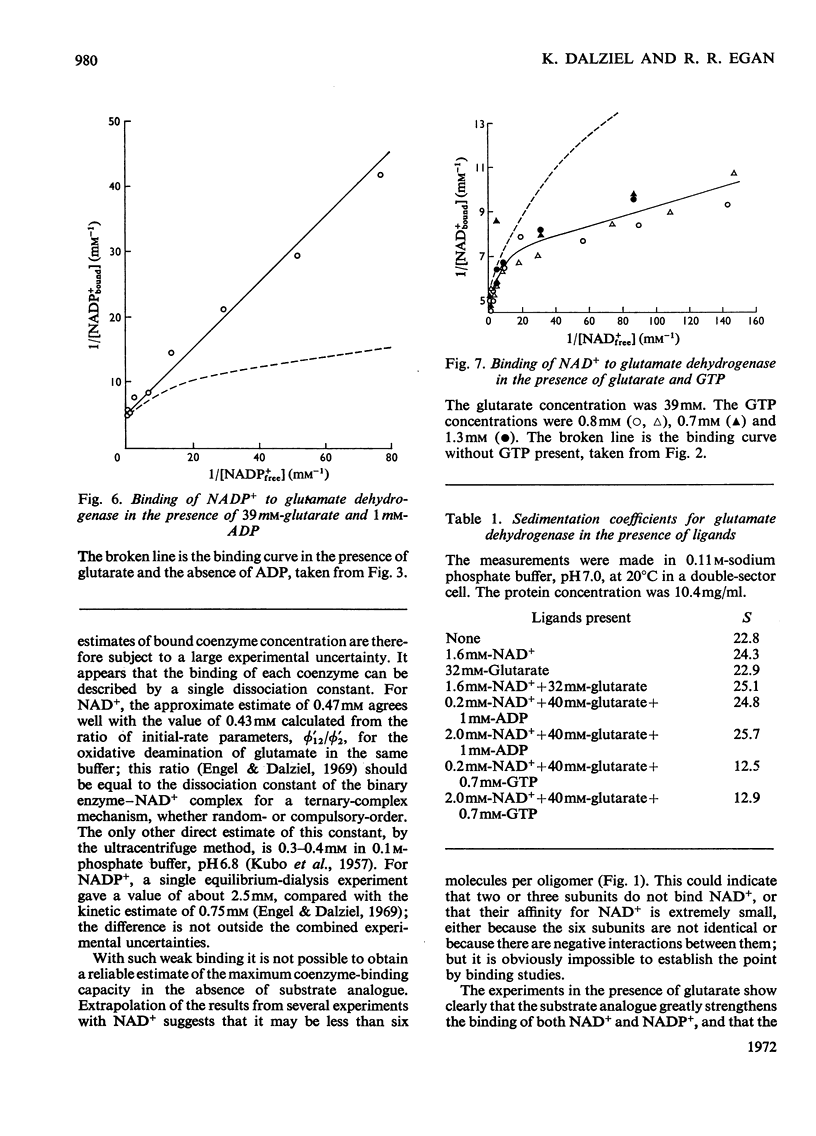
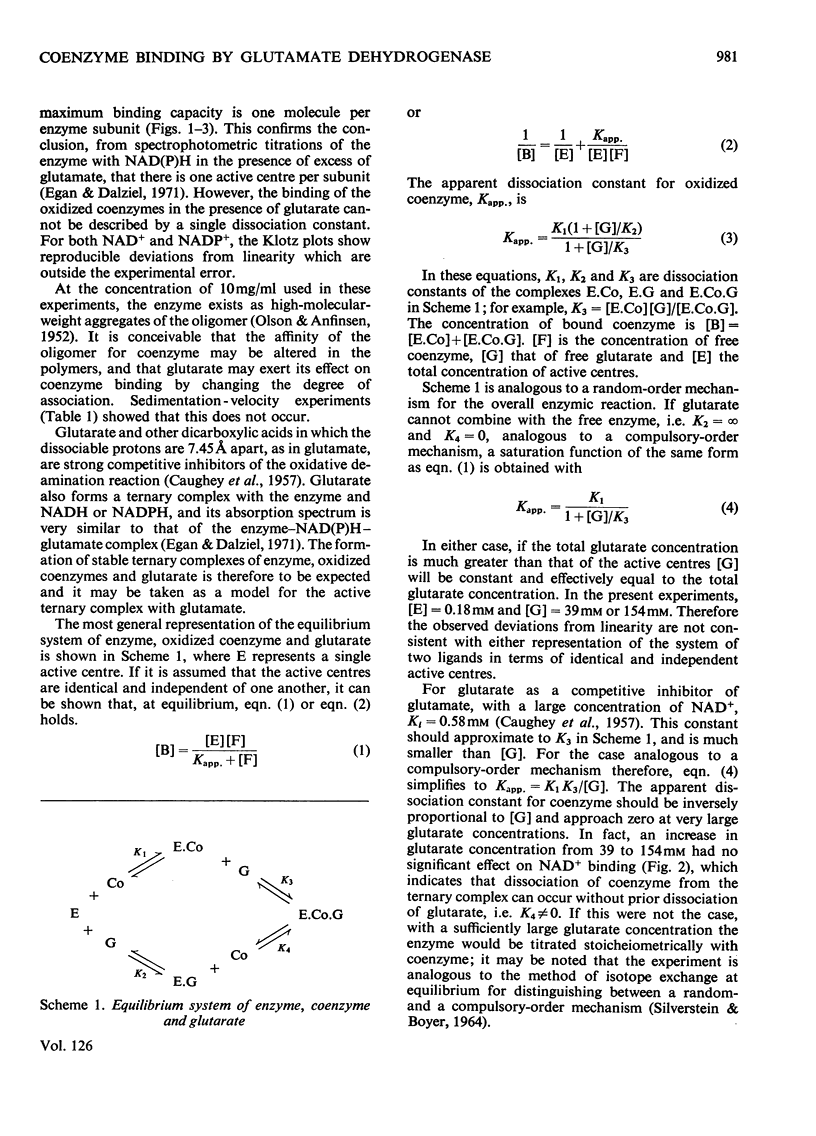
1. The binding of NAD+ and NADP+ to glutamate dehydrogenase has been studied in sodium phosphate buffer, pH7.0, by equilibrium dialysis. Approximate values for the dissociation constants are 0.47 and 2.5mm respectively. For NAD+ the value agrees with that estimated from initial-rate results. 2. In the presence of the substrate analogue glutarate both coenzymes are bound more firmly, and there is one active centre per enzyme subunit. The binding results cannot be described in terms of independent and identical active centres, and binding is stronger at low coenzyme concentrations than at high concentrations. Either the six subunits of the oligomer are not identical or there are negative interactions between them in the binding of coenzymes in ternary complexes with glutarate. The latter explanation is favoured. 3. The binding studies support the conclusions drawn from earlier kinetic studies of the glutamate reaction. 4. ADP and GTP respectively decrease and increase the affinity of the enzyme for NAD+ and NADP+, in both the presence and absence of glutarate. The negative binding interactions in the presence of glutarate are abolished by ADP, which decreases the affinity for the coenzymes at low concentrations of the latter. 5. In the presence of glutarate, GTP and NAD+ or NADP+, the association of enzyme oligomers is prevented, and the solubility of the enzyme is decreased; the complex of enzyme and ligands readily crystallizes. 6. The results are discussed in relation to earlier kinetic studies.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bayley P. M., Radda G. K. Conformational changes and the regulation of glutamate-dehydrogenase activity. Biochem J. 1966 Jan;98(1):105–111. doi: 10.1042/bj0980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAUGHEY W. S., HELLERMAN L., SMILEY J. D. L-glutamic acid dehydrogenase; structural requirements for substrate competition; effect of thyroxine. J Biol Chem. 1957 Jan;224(1):591–607. [PubMed] [Google Scholar]
- Cassman M., Schachman H. K. Sedimentation equilibrium studies on glutamic dehydrogenase. Biochemistry. 1971 Mar 16;10(6):1015–1024. doi: 10.1021/bi00782a013. [DOI] [PubMed] [Google Scholar]
- Conway A., Koshland D. E., Jr Negative cooperativity in enzyme action. The binding of diphosphopyridine nucleotide to glyceraldehyde 3-phosphate dehydrogenase. Biochemistry. 1968 Nov;7(11):4011–4023. doi: 10.1021/bi00851a031. [DOI] [PubMed] [Google Scholar]
- Cross D. G., Fisher H. F. The mechanism of glutamate dehydrogenase reaction. 3. The binding of ligands at multiple subsites and resulting kinetic effects. J Biol Chem. 1970 May 25;245(10):2612–2621. [PubMed] [Google Scholar]
- DALZIEL K. Possible magnitude of inhibition of coenzyme-substrate reactions by competitive inhibitors in coenzyme preparations. Nature. 1962 Jul 28;195:384–385. doi: 10.1038/195384a0. [DOI] [PubMed] [Google Scholar]
- Dalziel Keith, Engel Paul C. Antagonistic homotropic interactions as a possible explanation of coenzyme activation of glutamate dehydrogenase. FEBS Lett. 1968 Oct;1(5):349–352. doi: 10.1016/0014-5793(68)80153-x. [DOI] [PubMed] [Google Scholar]
- Di Prisco G. Tyrosyl and lysyl residues involved in the reactivity of catalytic and regulatory sites of crystalline beef liver glutamate dehydrogenase. Biochemistry. 1971 Feb 16;10(4):585–589. doi: 10.1021/bi00780a007. [DOI] [PubMed] [Google Scholar]
- Doddgh, Radda G. K. 1-Anilinonaphthalene-8-sulphonate, a fluorescent conformational probe for glutamate dehydrogenase. Biochem J. 1969 Sep;114(2):407–417. doi: 10.1042/bj1140407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan R. R., Dalziel K. Active centre equivalent weight of glutamate dehydrogenase from dry weight determinations and spectrophotometric titrations of abortive complexes. Biochim Biophys Acta. 1971 Oct;250(1):47–50. doi: 10.1016/0005-2744(71)90118-5. [DOI] [PubMed] [Google Scholar]
- Engel P. C., Dalziel K. Kinetic studies of glutamate dehydrogenase with glutamate and norvaline as substrates. Coenzyme activation and negative homotropic interactions in allosteric enzymes. Biochem J. 1969 Dec;115(4):621–631. doi: 10.1042/bj1150621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel P. C., Dalziel K. The equilibrium constants of the glutamate dehydrogenase systems. Biochem J. 1967 Nov;105(2):691–695. doi: 10.1042/bj1050691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRIEDEN C. GLUTAMATE DEHYDROGENASE. V. THE RELATION OF ENZYME STRUCTURE TO THE CATALYTIC FUNCTION. J Biol Chem. 1963 Oct;238:3286–3299. [PubMed] [Google Scholar]
- FRIEDEN C. Glutamic dehydrogenase. I. The effect of coenzyme on the sedimentation velocity and kinetic behavior. J Biol Chem. 1959 Apr;234(4):809–814. [PubMed] [Google Scholar]
- FRIEDEN C. Glutamic dehydrogenase. III. The order of substrate addition in the enzymatic reaction. J Biol Chem. 1959 Nov;234:2891–2896. [PubMed] [Google Scholar]
- Iwatsubo M., Pantaloni D. Régulation de l'activité de la glutamate déshydrogènase par les effecteurs GTP et ADP: étude par "stopped flow". Bull Soc Chim Biol (Paris) 1967 Dec 18;49(11):1563–1572. [PubMed] [Google Scholar]
- Koshland D. E., Jr, Némethy G., Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966 Jan;5(1):365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- MONOD J., WYMAN J., CHANGEUX J. P. ON THE NATURE OF ALLOSTERIC TRANSITIONS: A PLAUSIBLE MODEL. J Mol Biol. 1965 May;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- OLSON J. A., ANFINSEN C. B. Kinetic and equilibrium studies on crystalline 1-glutamic acid dehydrogenase. J Biol Chem. 1953 Jun;202(2):841–856. [PubMed] [Google Scholar]
- OLSON J. A., ANFINSEN C. B. The crystallization and characterization of L-glutamic acid dehydrogenase. J Biol Chem. 1952 May;197(1):67–79. [PubMed] [Google Scholar]
- Pantaloni D., Dessen P. Glutamate déshydrogénase. Fixations des coenzymes NAD et NADP et d'autres nucléotides dérivés de l'adénosine-5'-phosphate. Eur J Biochem. 1969 Dec;11(3):510–519. doi: 10.1111/j.1432-1033.1969.tb00803.x. [DOI] [PubMed] [Google Scholar]
- Pantaloni D., Iwatsubo M. Fixation d'ADP, NADH et NADPH sur la glutamatedéshydrogénase, déterminée par spectrophotométrie. Biochim Biophys Acta. 1967 Jan 11;132(1):217–220. doi: 10.1016/0005-2744(67)90217-3. [DOI] [PubMed] [Google Scholar]
- SILVERSTEIN E., BOYER P. D. EQUILIBRIUM REACTION RATES AND THE MECHANISMS OF LIVER AND YEAST ALCOHOL DEHYDROGENASE. J Biol Chem. 1964 Nov;239:3908–3914. [PubMed] [Google Scholar]
- Smith E. L., Landon M., Piszkiewicz D., Brattin W. J., Langley T. J., Melamed M. D. Bovine liver glutamate dehydrogenase: tentative amino acid sequence; identification of a reactive lysine; nitration of a specific tyrosine and loss of allosteric inhibition by guanosine triphosphate. Proc Natl Acad Sci U S A. 1970 Oct;67(2):724–730. doi: 10.1073/pnas.67.2.724. [DOI] [PMC free article] [PubMed] [Google Scholar]


