Abstract
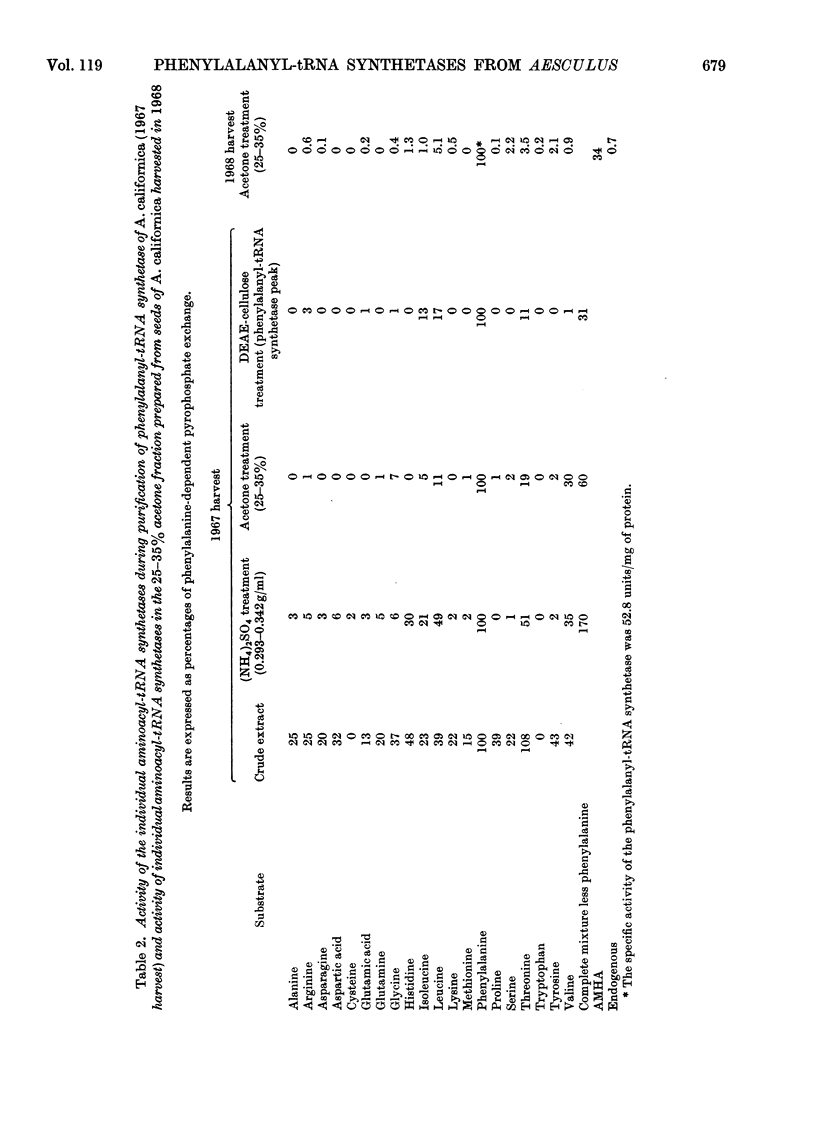
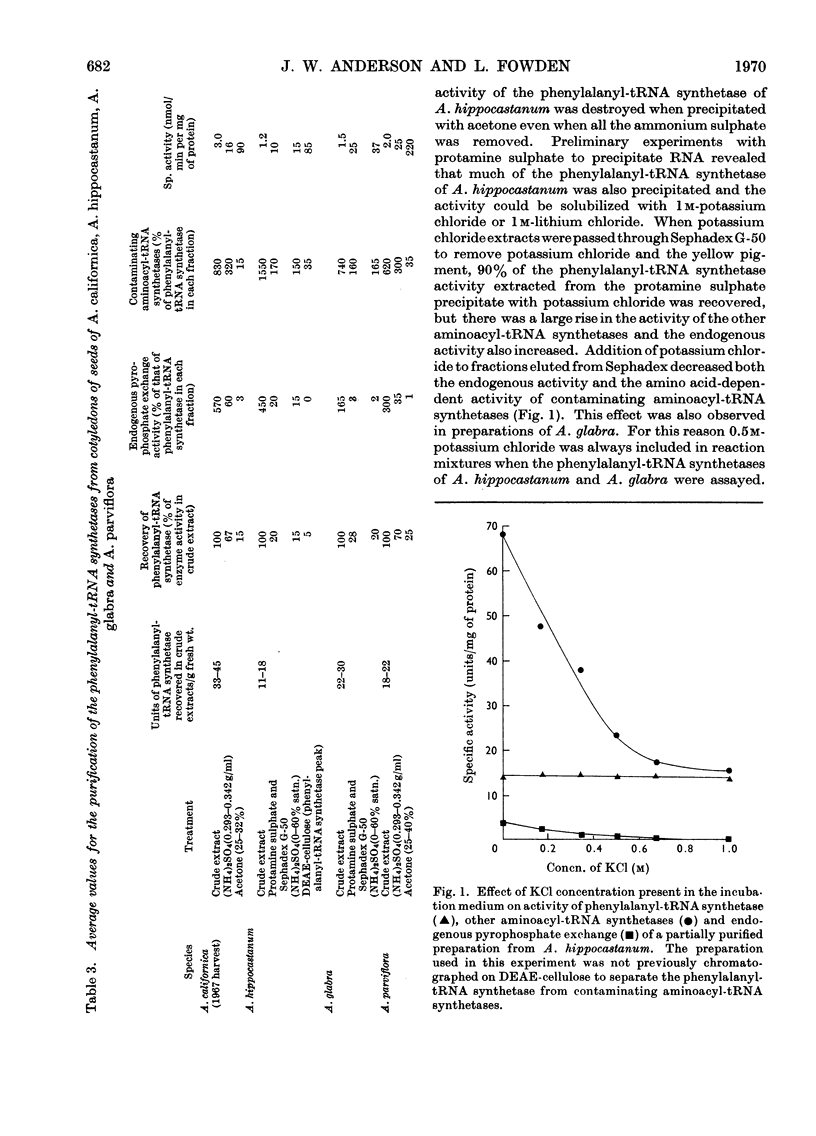
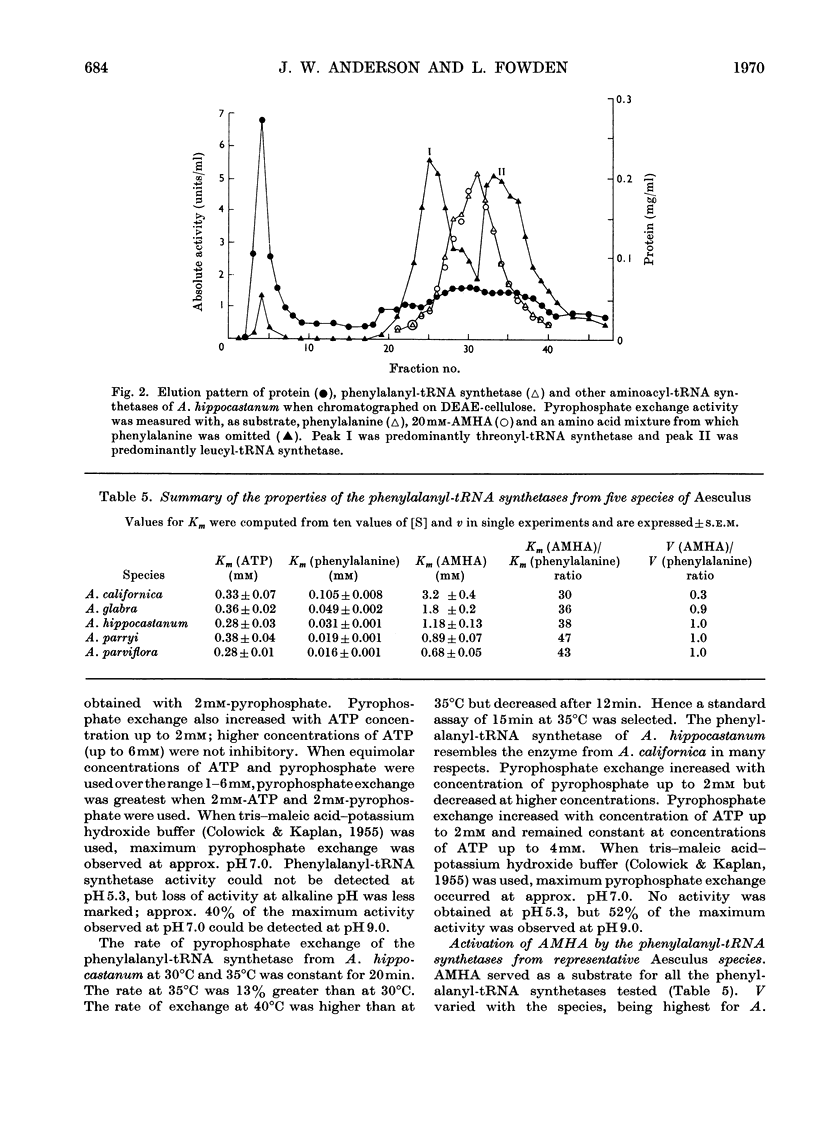
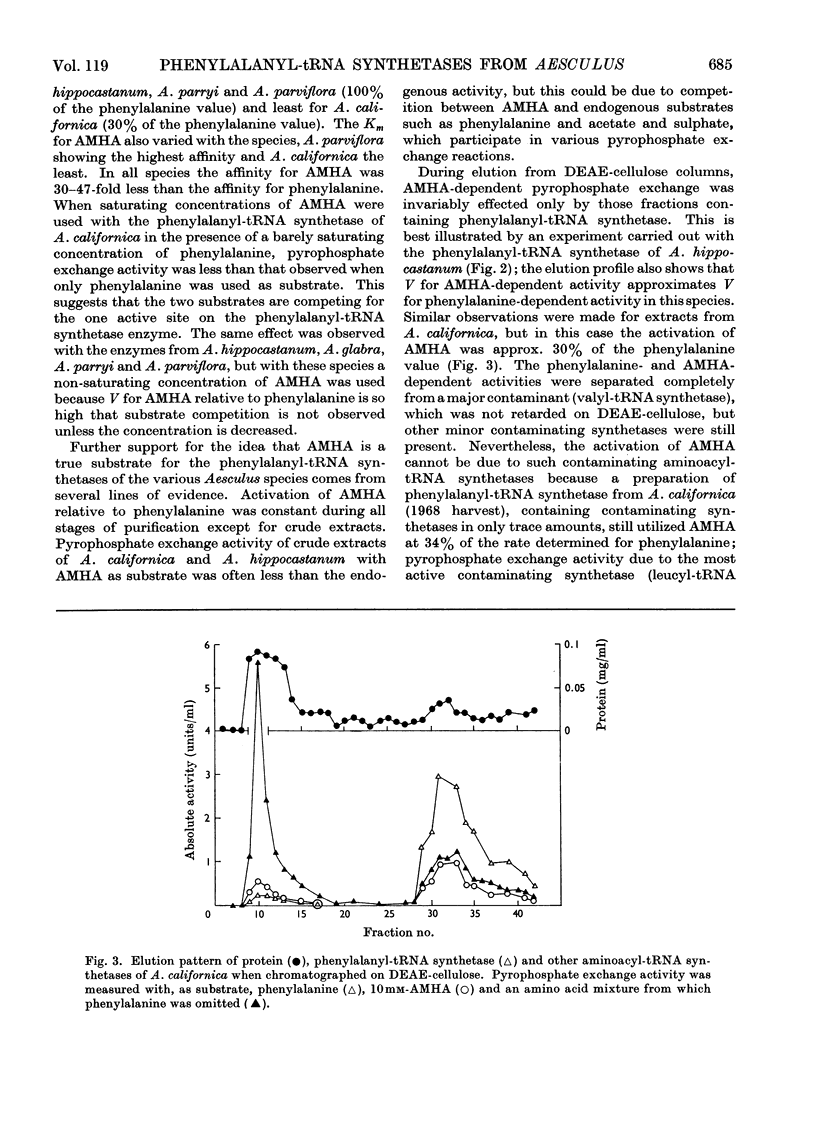
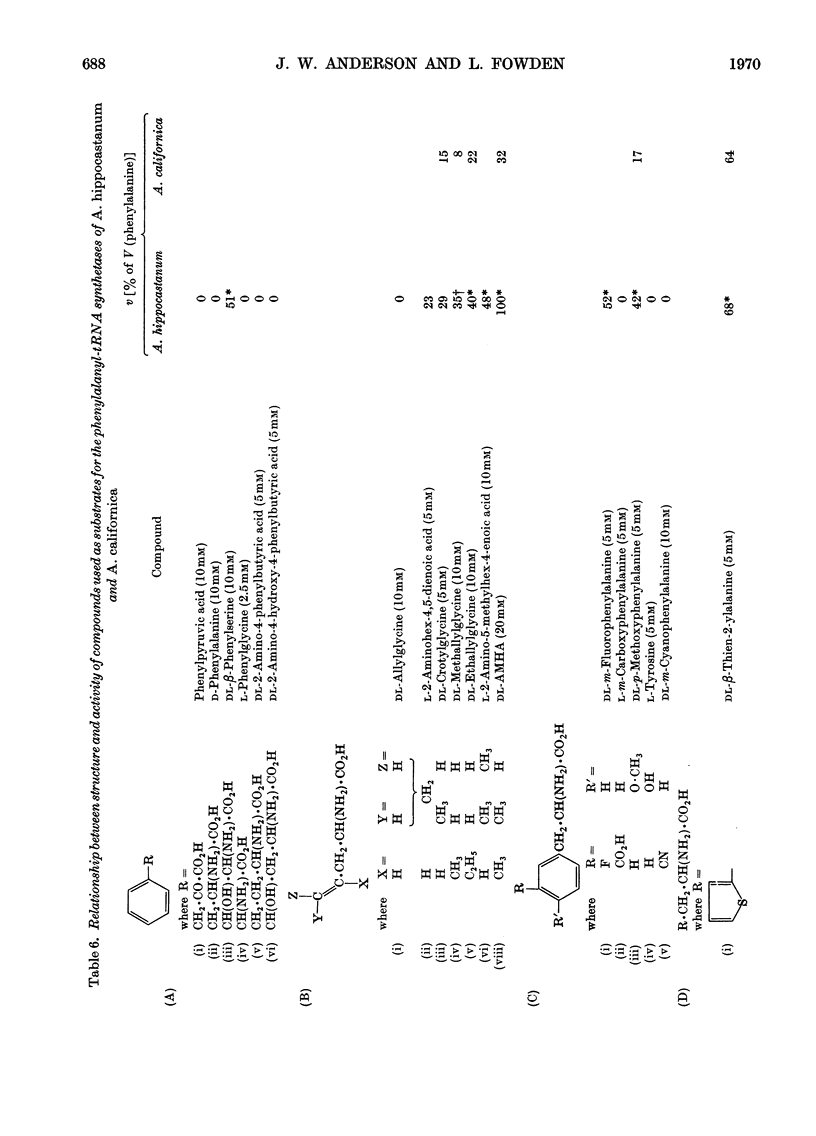
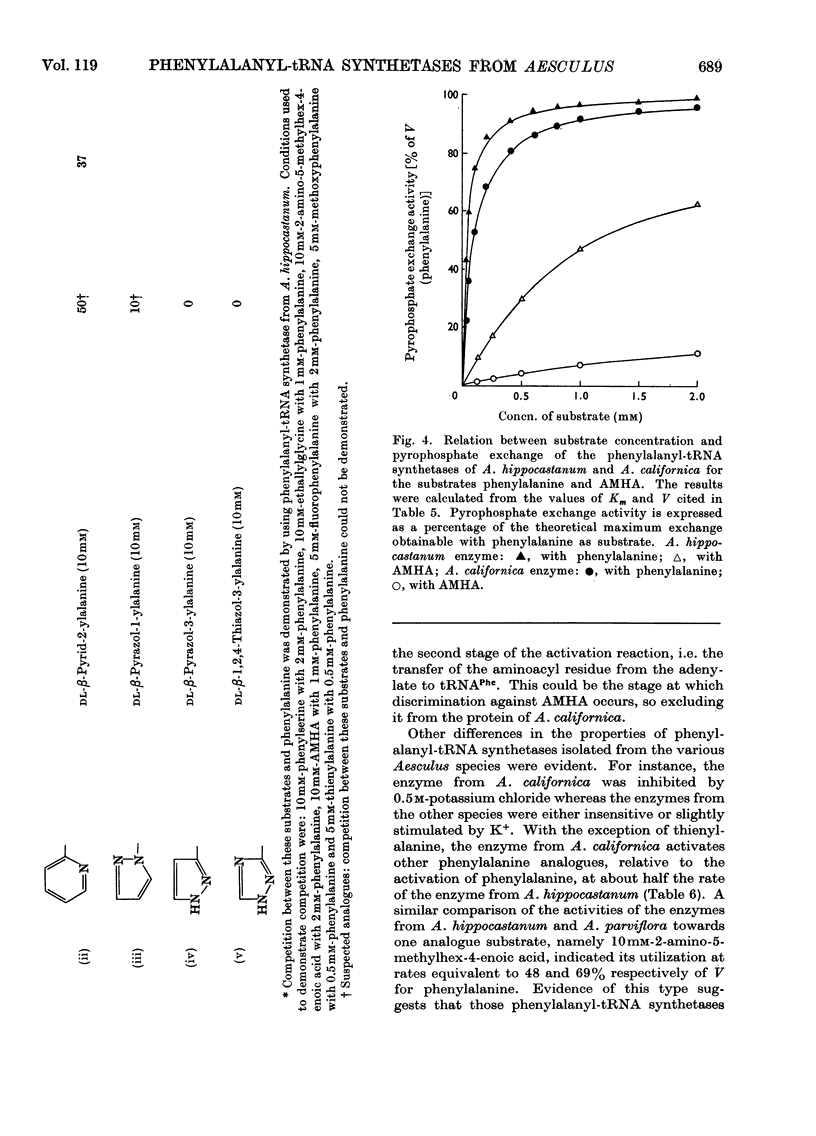
1. Phenylalanyl-tRNA synthetases have been partially purified from cotyledons of seeds of Aesculus californica, which contains 2-amino-4-methylhex-4-enoic acid, and from four other species of Aesculus that do not contain this amino acid. The A. californica preparation was free from other aminoacyl-tRNA synthetases, and the contaminating synthetase activity in preparations from A. hippocastanum was decreased to acceptable limits by conducting assays of pyrophosphate exchange activity in 0.5m-potassium chloride. 2. The phenylalanyl-tRNA synthetase from each species activated 2-amino-4-methylhex-4-enoic acid with Km 30–40 times that for phenylalanine. The maximum velocity for 2-amino-4-methylhex-4-enoic acid was only 30% of that for phenylalanine with the A. californica enzyme, but the maximum velocities for the two substrates were identical for the other four species. 3. 2-Amino-4-methylhex-4-enoic acid was not found in the protein of A. californica, so discrimination against this amino acid probably occurs in the step of transfer to tRNA, though subcellular localization, or subsequent steps of protein synthesis could be involved. 4. Crotylglycine, methallylglycine, ethallylglycine, 2-aminohex-4,5-dienoic acid, 2-amino-5-methylhex-4-enoic acid, 2-amino-4-methylhex-4-enoic acid, β-(thien-2-yl)alanine, β-(pyrazol-1-yl)alanine, phenylserine and m-fluorophenylalanine were substrates for pyrophosphate exchange catalysed by the phenylalanyl-tRNA synthetases of A. californica or A. hippocastanum. Allylglycine, phenylglycine and 2-amino-4-phenylbutyric acid were inactive.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson J. W., Rowan K. S. The extraction and assay of aminoacyl-transfer-ribonucleic acid synthetases of tobacco leaf. Biochem J. 1966 Oct;101(1):9–14. doi: 10.1042/bj1010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIE E. W., KONINGSBERGER V. V., LIPMANN F. The isolation of a tryptophan-activating enzyme from pancreas. Arch Biochem Biophys. 1956 Nov;65(1):21–38. doi: 10.1016/0003-9861(56)90173-4. [DOI] [PubMed] [Google Scholar]
- ELLMAN G. L. The biuret reaction: changes in the ultraviolet absorption spectra and its application to the determination of peptide bonds. Anal Biochem. 1962 Jan;3:40–48. doi: 10.1016/0003-2697(62)90042-8. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Legocki A. B., Pawelkiewicz J. Amino acid-activating enzymes in yellow lupin seeds, and purification of leucyl-sRNA synthetase. Acta Biochim Pol. 1967;14(3):313–322. [PubMed] [Google Scholar]
- Peterson P. J. Amino acid selection in protein biosynthesis. Biol Rev Camb Philos Soc. 1967 Nov;42(4):552–613. doi: 10.1111/j.1469-185x.1967.tb01530.x. [DOI] [PubMed] [Google Scholar]
- Peterson P. J., Fowden L. Purification, properties and comparative specificities of the enzyme prolyl-transfer ribonucleic acid synthetase from Phaseolus aureus and Polygonatum multiflorum. Biochem J. 1965 Oct;97(1):112–124. doi: 10.1042/bj0970112. [DOI] [PMC free article] [PubMed] [Google Scholar]


