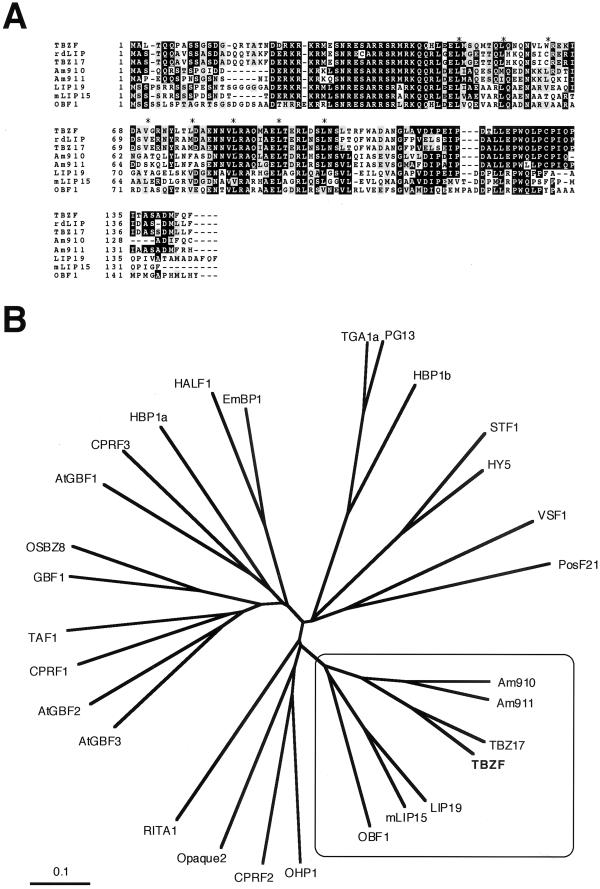
Figure 1.
A, Amino acid sequence alignment of TBZF and LIP19 subfamily proteins. Identical residues are highlighted in black and similar residues in gray. Dashes were introduced to maximize the alignment. A basic region and the positions of heptad Leu residues are indicated by hooked line and asterisks, respectively. TBZF (this paper), rdLIP (accession no. AB005187; Ito et al., 1999), TBZ17 (accession no. D63951; Kusano et al., 1998), Am910 (accession no. T17108), Am911 (accession no. T17110; Martínez-García et al., 1998), LIP19 (accession no. X57325; Aguan et al., 1993), mLIP15 (accession no. D26563; Kusano et al., 1995), and OBF1 (accession no. X62745; Singh et al., 1990) are used for multi-alignment. B, Phylogenic tree of plant bZIP proteins. Trees were built using Clustal X program (Thompson et al., 1997) and visualized by a TreeView program (Page, 1996). Accession nos. are as follows: AtGBF1–3 (accession nos. X63894–X63896), CPRF1–3 (accession nos. X58575–X58577), EmBP1 (accession no. P25032), GBF1 (accession no. U10270), HALF1 (accession no. D64051; 1996), HBP1a (accession no. X56781), HBP1b (accession no. X56782), HY5 (accession no. AB005456), OHP1 (accession no. L00623), Opaque 2 (accession nos. X15544 and M29411), OSBZ8 (accession no. U42208), PG13 (accession no. M62855), PosF21 (accession no. X61031), RITA1 (accession no. L34551), STF1 (accession no. L28003), TAF1 (accession no. X60363), TGA1a (accession no. S05452), TGA1b (accession no. S05453), and VSF1 (accession no. X73635). LIP19 subfamily was squared and TBZF was highlighted by bold type. The bar corresponds to 0.1 Jukes-Cantor substitutions per nucleotide.