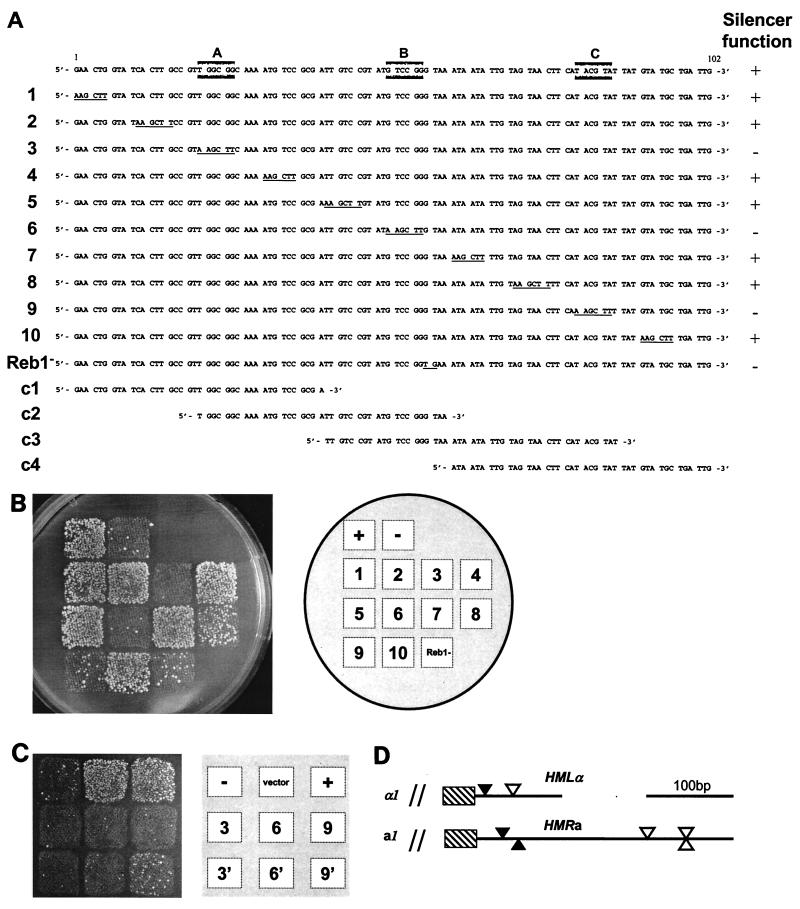
FIG. 1.
Three discrete sites were required for silencer function. (A) DNA sequence of the HMLα silencer and mutations generated. The top line represents the wild-type sequence, and lines 1 to 10 represent mutations generated in the silencer. Changed bases are underlined. Reb1− shows the 2-bp substitution in the Reb1p recognition sequence. Lines c1 to c4 represent the DNA fragments used for competition in the gel mobility shift experiment. A, B, and C (shaded boxes) indicate the sequences defined to be essential for silencer function. Silencer function in the mating assay is indicated on the right. (B) Mating assay for silencer function. A tester mater strain (MATα WM52V4) was mated to a MATa strain (CK213-4C) containing different plasmids. These plasmids contained the HMLα locus either lacking a silencer (p291; −) or containing a wild-type silencer (p413; +). Mutant silencers were as indicated in panel A. Diploids were selected on synthetic dextrose plates. (C) Introduction of ORC binding sites did not improve the silencer function of A and B box mutations. A mating assay was performed as described in panel B. Squares 3, 6, and 9 are the same silencer mutations indicated in panel A, and 3′, 6′, and 9′ indicate that the adjacent ORC binding site was included on the plasmid. A plasmid without an insert is denoted as the vector. (D) Schematic drawing of the HMLα and HMRa silencers. Sequence analysis with MatInspector software revealed the presence of both A boxes (solid arrowhead) and B boxes (Reb1 binding-sites) (open arrowhead) at both the HMLα and HMRa silencers.