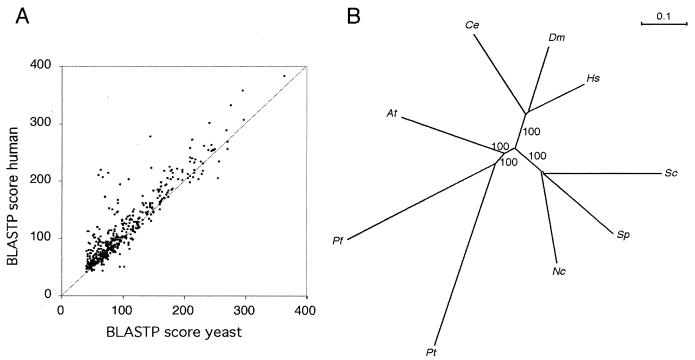
FIG. 5.
Comparative analysis. (A) BLASTP scores for the 447 conserved partial ORFs are shown as a scatter plot, with the best score against the yeast proteome on the abcissa and the best score against the human proteome on the ordinate axis. (B) Molecular phylogeny by using combined protein data. Unrooted distance matrix neighbor-joining tree obtained for nine species with a set of 38 partial proteins comprising 4,037 informative amino acids. The bootstrap values for 100 replicates were 100 at all nodes except for the two very closely spaced nodes connecting the three fungal species. At, Arabidopsis thaliana; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Hs, Homo sapiens; Nc, Neurospora crassa; Pf, Plasmodium falciparum; Pt, Paramecium tetraurelia; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe.