Figure 4.
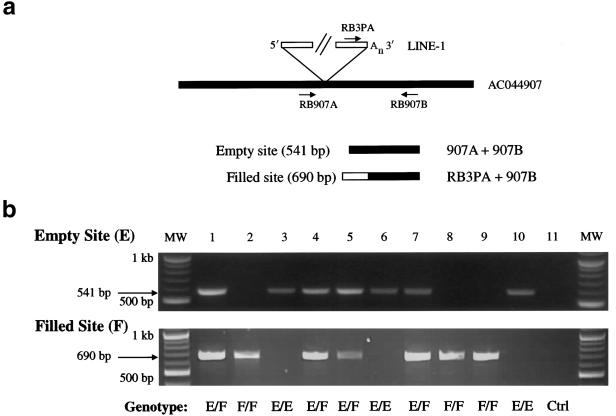
Dimorphic Ta-1d L1s, identified by 5′ ATLAS. a, Independent assay to assess the dimorphic status of Ta-1d L1s. PCR typing assays were developed to confirm the presence (filled)/absence (empty) status of Ta-1d L1s identified in the 5′ ATLAS reactions shown in panel b. Empty sites, lacking the L1 insertion, can be amplified with primers flanking the insertion site (e.g., RB907A and RB907B). Filled sites, containing the L1 insertion, can be amplified with a Ta-subfamily–specific L1 3′ UTR primer and a 3′ flanking primer (e.g., RB3PA and RB907B). b, Results of PCR typing experiments. Top, Amplification of empty site through PCR typing by use of the RB907A and RB907B primers. Bottom, Amplification of filled site through PCR typing by use of the RB3PA and RB907B primers. Note that individual DNA samples (1–10) can be homozygous for the presence of the L1 (F/F; lanes 2, 8, and 9), heterozygous (EF; lanes 1, 4, 5, and 7), or homozygous for the absence of the L1 (E/E; lanes 3, 6, and 10). The 11th reaction (Ctrl) is a control PCR with genomic DNA omitted. MW = molecular weight markers.