Abstract
Dyschromatosis symmetrica hereditaria (DSH) is a hereditary skin disease characterized by the presence of hyperpigmented and hypopigmented macules on extremities and face. The gene, or even its chromosomal location, for DSH has not yet been identified. In this study, two Chinese families with DSH were identified and subjected to a genomewide screen for linkage analysis. Two-point linkage analysis for pedigree A (maximum LOD score [Zmax] = 7.28 at recombination fraction [θ] = 0.00) and pedigree B (Zmax = 2.41 at θ = 0.00) mapped the locus for DSH in the two families to chromosome 6q. Subsequent multipoint analysis of the two families also provided additional support for the DSH gene being located within the region 6q24.2-q25.2, with Zmax = 10.64. Haplotype analysis confined the locus within an interval of 10.2 Mbp, flanked by markers D6S1703 and D6S1708. The two families had no identical haplotype within the defined region, which suggests that the two families were different in origin. Further work on identification of the gene for DSH will open new avenues to exploration of the genetics of pigmentation.
Dyschromatosis symmetrica hereditaria (DSH [MIM 127400]), also called “symmetric dyschromatosis of the extremities” or “reticulate acropigmentation of Dohi,” was first reported by Matsumoto (1923) and named by Toyama as a clinical entity (Toyama 1929). DSH generally shows an autosomal dominant pattern of inheritance, with high penetrance (Oyama et al. 1999). DSH can be characterized by hyperpigmented and hypopigmented macules, on face and dorsal aspects of extremities, that appear in infancy or early childhood (Patrizi et al. 1994; Ostlere et al. 1995; Danese et al. 1997; Oyama et al. 1999; Ohtoshi et al. 2001). The skin lesions commonly stop spreading before adolescence but last for life. These abnormalities are otherwise asymptomatic and do not affect general health. Histologically, melanin pigmentation was increased in the basal cells of hyperpigmented lesions, but the numbers of melanocytes were decreased in hypopigmented macules (Oyama et al. 1999). Electron microscopic findings in a hypermelanotic area showed an increased number of melanocytes with high metabolic activity. In the hypomelanotic areas, the melanocytes were morphologically abnormal, with melanosomes at the early stages of development (Danese et al. 1997; Ohtoshi et al. 2001).
In Asia, DSH occurs predominantly among Japanese and Korean individuals (Danese et al. 1997; Oyama et al. 1999) but has been reported in other Asian countries to a lesser extent (Tan and Tay 1997; Hemanthkumar and Thappa 1997). Because DSH is a relatively common phenotype in the white population, particularly in redheads, in the West, there is great concern about DSH. So far, many clinical and morphological investigations have been reported, but the DSH gene—including the pathogenesis—has not yet been identified (Kono et al. 2000). In an effort to localize the gene for DSH, we embarked on a genomewide search and found significant evidence for linkage of the gene responsible for DSH to chromosome 6q24.2-25.2.
Two families from the Henan and Yunnan provinces of China with typical features of DSH were recruited for this work. Both pedigrees with DSH showed an autosomal dominant inheritance pattern (fig. 1) and were ascertained by the Huashan Hospital of Fudan University, the First Affiliated Hospital of Zhengzhou University, and the Shanghai Ninth People’s Hospital of Shanghai Second Medical University. The affected individuals had symmetrical cutaneous hypopigmented and hyperpigmented macules, ranging in size from pea-sized to covering the backs of fingers and the backs of hands. The similar distributions could be found on feet and even on trunks (fig. 2). Biopsies of hyperpigmented and hypopigmented macules on the tops of the feet revealed basal melanosis and hypomelanosis, which were similar to the reports on Japanese subjects. Nevertheless, none of the affected members in either family was found to have skin cancer.
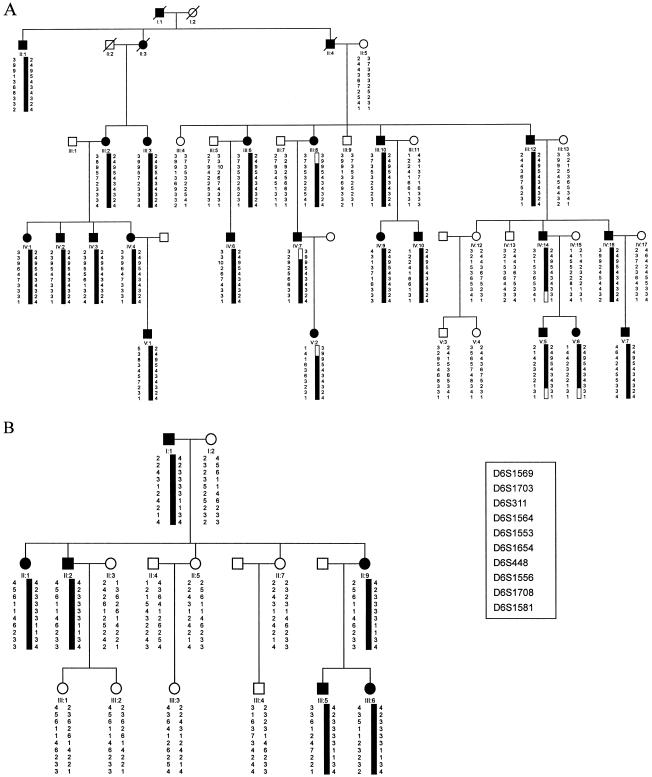
Figure 1.
Pedigree structure and haplotypes of family A (A) and family B (B). Marker order was determined from the Généthon sex-averaged genetic map, the CHLC sex-averaged genetic map, and the Genome Database. Open symbols indicate unaffected individuals, blackened symbols indicate affected individuals, squares indicate men, and circles indicate women. Blackened bars indicate the chromosome region shared by affected members of the pedigree.
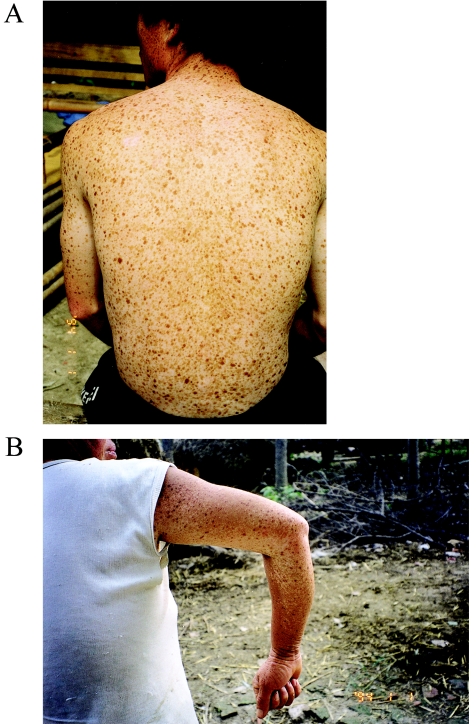
Figure 2.
A mixture of hypopigmented and hyperpigmented macules of various sizes on the trunk and arms of DSH-affected individuals in pedigree A (A) and pedigree B (B), respectively.
This research has been approved by the ethical review committees of the appropriate institutions. In total, 50 family members participated in this study, after having given informed consent. Samples of peripheral-blood DNA were taken from all available family members, and DNA was prepared by means of our standard procedure (Gao et al. 2001; Liu et al. 2001). In linkage analysis, the samples were analyzed individually, rather than by the DNA-pooling method usually used in our other work (Liu et al. 2001).
We therefore performed a genomewide scan in these two Chinese families with DSH to determine the chromosomal regions linked to DSH, using 382 polymorphic microsatellite markers covering 22 autosomes, with an average marker density of 10 cM, according to the Généthon linkage map. The markers used were from the ABI Prism Linkage Mapping Set (version 2.0) and the Généthon Human Linkage Map (Dib et al. 1996). Marker order and intermarker distances were obtained from linkage map of the Cooperative Human Linkage Center (CHLC). Semiautomated fluorescence genotyping was adopted to type the microsatellite markers (Yang et al. 2000). PCR was performed under the conditions recommended by the manufacturer (PE Biosystems). PCR products were mixed with a loading cocktail that contained formamide, Gs-400HD ROX standards (PE Biosystems), and loading dye. The product was loaded onto 6% acrylamide gel and run in an ABI 377 Prism DNA Sequencer (Perkin Elmer). The data were analyzed by ABI GENESCAN 3.1 and ABI GENOTYPER 2.1 software (PE Biosystems).
A two-point LOD score was calculated, by use of the MLINK routine of the FASTLINK software package version 5.1 (Lathrop and Lalouel 1984), under an assumed genetic model: autosomal dominant, a disease-allele frequency of 0.0001, evenly shared allele frequency, zero phenocopy rate, no sex difference, and full penetrance (Kono et al. 2000). Multipoint analysis was performed with the FASTMAP program (Curtis and Gurling 1993).
Initial evidence of linkage to DSH was obtained in pedigree A at marker D6S1581, from which a LOD score of 2.24 was obtained at θ=0.05. In further refinement study with high-density markers in the same region, we confirmed the finding with the highest LOD score (Zmax) of 7.28 (recombination fraction [θ] = 0.00) at marker D6S1654 (table 1). Conversely, no significant linkage with markers on other chromosomal regions was found. Subsequent multipoint analysis carried out with FASTMAP also provided additional support for the localization of DSH locus to chromosome 6q24.2-q25.2, with Zmax=7.84. In the next study of pedigree B, using the markers that generated positive results for pedigree A, we obtained Zmax=2.41 at θ=0.00 at marker D6S448, which is positioned ∼2.9 cM away from D6S1654 (table 1). The multipoint analysis produced Zmax=3.09 in the same region. Obviously, the results suggest that the gene locus responsible for the disorder in family B is also localized in chromosome 6q, in light of the restricted size of the family. The combined multipoint LOD score for both families is 10.64 around marker D6S1553.
Table 1.
Two-Point LOD Scores Obtained from Linkage Analysis between DSH Locus and Chromosome 6q Markers in Two Pedigrees
| LOD Score at θ = |
|||||||
| Locus and Pedigree | .0 | .01 | .05 | .1 | .2 | .3 | .4 |
| D6S1569: | |||||||
| Ped A | −∞ | −2.22 | −.45 | .03 | .13 | .04 | .03 |
| Ped B | 1.51 | 1.47 | 1.35 | 1.19 | .83 | .46 | .13 |
| D6S1703: | |||||||
| Ped A | −∞ | 3.74 | 4.06 | 3.87 | 3.12 | 2.13 | 1.02 |
| Ped B | 1.20 | 1.19 | 1.12 | 1.02 | .82 | .58 | .32 |
| D6S311: | |||||||
| Ped A | 3.59 | 3.53 | 3.29 | 2.98 | 2.31 | 1.59 | .83 |
| Ped B | 2.11 | 2.07 | 1.93 | 1.74 | 1.33 | .87 | .39 |
| D6S1564: | |||||||
| Ped A | 4.63 | 4.55 | 4.24 | 3.83 | 2.94 | 1.97 | .91 |
| Ped B | .87 | .85 | .78 | .70 | .52 | .35 | .17 |
| D6S1553: | |||||||
| Ped A | 6.98 | 6.87 | 6.41 | 5.81 | 4.52 | 3.08 | 1.49 |
| Ped B | 2.11 | 2.07 | 1.93 | 1.74 | 1.33 | .87 | .39 |
| D6S1654: | |||||||
| Ped A | 7.28 | 7.16 | 6.69 | 6.06 | 4.72 | 3.22 | 1.57 |
| Ped B | 2.11 | 2.07 | 1.93 | 1.74 | 1.33 | .87 | .39 |
| D6S448: | |||||||
| Ped A | 6.39 | 6.28 | 5.82 | 5.23 | 3.94 | 2.55 | 1.12 |
| Ped B | 2.41 | 2.37 | 2.21 | 2.00 | 1.54 | 1.02 | .47 |
| D6S1556: | |||||||
| Ped A | 3.29 | 3.23 | 3.01 | 2.72 | 2.11 | 1.45 | .75 |
| Ped B | 2.07 | 2.03 | 1.87 | 1.67 | 1.24 | .78 | .32 |
| D6S1708: | |||||||
| Ped A | −∞ | 3.24 | 3.56 | 3.38 | 2.70 | 1.87 | .92 |
| Ped B | 1.20 | 1.18 | 1.09 | .98 | .72 | .44 | .15 |
| D6S1581: | |||||||
| Ped A | −∞ | 1.15 | 2.24 | 2.43 | 2.14 | 1.52 | .72 |
| Ped B | 1.20 | 1.19 | 1.12 | 1.02 | .82 | .58 | .32 |
Subsequent haplotypes were performed with Cyrillic version 2.1 (Cherwell Scientific) to confine interval of the linked region (fig. 1). The recombination events between the DSH phenotype and the markers that span the region of interest defined the smallest cosegregating region that included critical meiotic recombinants in pedigree A. Careful examination of the haplotypes confirmed that disease-associated alleles cosegregated with the phenotype of DSH in pedigree A. A recombination event in individual III-8 placed the disease locus proximal to D6S1703, since the affected individual did not inherit the disease-linked alleles of D6S1703 in family A. A recombination event in individual IV-14 placed the disease locus distal to D6S1708, since this affected individual did not share the disease-linked alleles of D6S1708 in family A. Hence, the maximal interval of linkage with DSH phenotype is bordered by D6S1703 (centromeric) and D6S1708 (telomeric) within a region of ∼10.2 Mbp, according to the last draft of the human genome sequence (Build 31).
The haplotype construction of pedigree B showed that the affected individuals shared a common allele, for the markers in the susceptibility region, that was not shared by the healthy individuals. This suggests that the gene responsible for the disorder in family B is also localized in chromosome 6q24.2-q25.2. However, the haplotypes in the two families contain the same contiguous markers but with different alleles. In this study, we seem to encounter the similar situation: that the two families may share the same disorder-causing gene but of different origin (Yang et al. 2000). However, the above results well demonstrate that at least one gene responsible for DSH, likely with different mutations, is located within the same region (Gao et al. 2001).
Clinical manifestations of DSH are dominated by hyperpigmented and hypopigmented macules of various sizes on the face and extremities, even on the trunk. Melanin pigmentation plays an important role in protecting skin against the damaging effects of ultraviolet rays. Although DSH is a disorder of pigmentation, no evidence has been found that DSH confers an increased risk of forming melanoma. In several cases, skin lesions were reported to become more pronounced after sun exposure, but there was no evidence revealed of photosensitivity (Satoh and Yoshida 1980).
Several human genetic diseases, such as LEOPARD syndrome (MIM 151100) and xeroderma pigmentation (XP), show some overlap with DSH. LEOPARD syndrome is an autosomal dominant inheritance characterized by multiple lentigines, electrocardiographic conduction abnormalities, ocular hypertelorism, pulmonic stenosis, abnormal genitalia, retardation of growth, and sensorineural deafness (Voron et al. 1976). Diffuse lentiginosis is a distinct characteristic of LEOPARD syndrome and similar to hyperpigmented macules of DSH. However, DSH has hypopigmentation macules and no common associated disorders. In addition, the gene for LEOPARD syndrome has previously been mapped to chromosome 12q24.1. Thus, the genetic basis of DSH is likely to differ from that of LEOPARD syndrome. With regard to XP, it is an autosomal recessive syndrome with clinical manifestations of excessive freckling, depigmentation, hyperpigmentation, skin aging, and a very high level of early and multiple skin cancers (Stary and Sarasin 2002). Although a mild type of XP also resembles DSH, DSH does not show the xerosis, atrophy, telangiectasia, or skin tumors usually observed in XP.
DSH is also likely to be genetically heterogeneous, since examples of an autosomal dominant (Oyama et al. 1999) and autosomal recessive (Urabe and Hori 1997; Alfadley et al. 2000) inheritance have been reported. In this work, we have mapped dominant DSH to the region within 10.2 Mbp of chromosome 6q24.2-q25.2, where there have been >60 possible genes identified. It is unfortunate that no gene or sequence in the region has been found to be homologous to any of >100 known loci that affect pigmentation in the mouse, according to the search of all currently available databases (Coat Color Genes) (Jackson 1997; Nakamura et al. 2002). However, several genes in this region—such as RGS17 (MIM 607191), AKAP12 (MIM 604698), and MAP3K7IP2 (MIM 605101), which play roles in signal pathways—can be considered as candidate genes for DSH. The products of RGS17, a member of the regulators of G protein–signaling (RGS) proteins that contain homologous core domains (RGS domains) of ∼120 amino acids, are important regulatory and structural components of G protein–coupled receptor complexes and are involved in modulating a variety of cell functions, such as proliferation, differentiation, response to neurotransmitters, membrane trafficking, and embryonic development (De Vries and Gist Farquhar 1999; Sierra et al. 2002). We are currently making efforts in identification of these candidate genes.
In conclusion, on the basis of this work, we clearly show that a major gene responsible for dominant DSH in the human genome has been first localized in the region of chromosome 6q24.2-q25.2. The characterization of the DSH gene will provide important clues to understand the molecular mechanism of pigmentation and increase ultimate hope for effectively interventional strategies.
Acknowledgments
This work was supported by grants from the national 973 and 863 Projects, the National Natural Science Foundation of China, K. C. Wang Education Foundation (Hong Kong), and Shanghai Municipal Commission for Science and Technology.
Electronic-Database Information
URLs for data presented herein are as follows:
- Coat Color Genes, http://www.cbc.umn.edu/ifpcs/micemut.htm
- Cooperative Human Linkage Center, http://gai.nci.nih.gov/CHLC/ (for marker maps)
- Généthon, http://www.genethon.fr/php/index.php (for extra markers for regions of interest)
- The Genome Database, http://www.gdb.org/ [Google Scholar]
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for DSH, LEOPARD syndrome, RGS17, AKAP12, and MAP3K7IP2)
References
- Alfadley A, Al Ajlan A, Hainau B, Pedersen K-T, Al Hoqail I (2000) Reticulate acropigmentation of Dohi: a case report of autosomal recessive inheritance. J Am Acad Derm 43:113–117 [DOI] [PubMed] [Google Scholar]
- Curtis D, Gurling H (1993) A procedure for combining two-point LOD scores into a summary multipoint map. Hum Hered 43:173–185 [DOI] [PubMed] [Google Scholar]
- Danese P, Zanca A, Bertazzoni MG (1997) Familial reticulate acropigmentation of Dohi. J Am Acad Dermatol 37:884–886 [DOI] [PubMed] [Google Scholar]
- De Vries L, Gist Farquhar M (1999) RGS proteins: more than just GAPs for heterotrimeric G proteins. Trends Cell Biol 9:138–144 [DOI] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- Gao B, Guo J, She C, Shu A, Yang M, Tan Z, Yang X, Guo S, Feng G, He L (2001) Mutations in IHH, encoding India hedgehog, cause brachydactyly type A-1. Nat Genet 28:386–389 [DOI] [PubMed] [Google Scholar]
- Jackson IJ (1997) Homologous pigmentation mutations in human, mouse and other model organisms. Hum Mol Genet 6:1613–1624 [DOI] [PubMed] [Google Scholar]
- Kono M, Miyamura Y, Matsunaga J, Tomita Y (2000) Exclusion of linkage between dyschromatosis symmetrica hereditaria and chromosome 9. J Dermatol Sci 22:88–95 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM (1984) Easy calculations of LOD scores and genetic risks on small computers. Am J Hum Genet 36:460–465 [PMC free article] [PubMed] [Google Scholar]
- Liu W, Wang H, Zhao S, Zhao W, Bai S, Zhao Y, Xu S, Wu C, Huang W, Chen Z, Feng G, He L (2001) The novel gene locus for agenesis of permanent teeth (He-Zhao deficiency) maps to chromosome 10q11.2. J Dent Res 80:1716–1720 [DOI] [PubMed] [Google Scholar]
- Matsumoto S (1923) Leucopathia punctata et reticularis symmetrica (in Japanese). Acta Dermatol 2:191–197 [Google Scholar]
- Nakamura M, Tobin DJ, Richards-Smith B, Sundberg JP, Paus R (2002) Mutant laboratory mice with abnormalities in pigmentation: annotated tables. J Dermatol Sci 28:1–33 [DOI] [PubMed] [Google Scholar]
- Ohtoshi E, Matsumura Y, Nishigori C, Toda KI, Horiguchi Y, Ikenaga M, Miyachi Y (2001) Useful applications of DNA repair tests for differential diagnosis of atypical dyschromatosis symmetrica hereditaria from xeroderma pigmentosum. Br J Dermatol 144:162–168 [DOI] [PubMed] [Google Scholar]
- Ostlere LS, Ratnavel RC, Lawlor F, Black MM, Griffiths WA (1995) Reticulate acropigmentation of Dohi. Clin Exp Dermatol 20:477–479 [DOI] [PubMed] [Google Scholar]
- Oyama M, Shimizu H, Ohata Y, Tajima S, Nishikawa T (1999) Dyschromatosis symmetrica hereditaria (reticulate acropigmentation of Dohi): report of a Japanese family with the condition and a literature review of 185 cases. Br J Dermatol 140:491–496 [DOI] [PubMed] [Google Scholar]
- Patrizi A, Manneschi V, Pini A, Baioni E, Ghetti P (1994) Dyschromatosis symmetrica hereditaria associated with idiopathic torsion dystonia: a case report. Acta Derm Venerol 74:135–137 [DOI] [PubMed] [Google Scholar]
- Satoh Y, Yoshida M (1980) Clinical and photobiological differences between dyschromatosis symmetrica hereditaria and xeroderma pigmentosum. J Dermatol 7:317–322 [DOI] [PubMed] [Google Scholar]
- Sierra DA, Gilbert DJ, Householder D, Grishin NV, Yu K, Ukidwe P, Barker SA, He W, Wensel TG, Otero G, Brown G, Copeland NG, Jenkins NA, Wilkie TM (2002) Evolution of the regulators of G-protein signaling multigene family in mouse and human. Genomics 79:177–185 [DOI] [PubMed] [Google Scholar]
- Stary A, Sarasin A (2002) The genetics of the hereditary xeroderma pigmentosum syndrome. Biochimie 84:49–60 [DOI] [PubMed] [Google Scholar]
- Tan HH, Tay YK (1997) Neurofibromatosis and reticulate acropigmentation of Dohi: a case report. Pediatr Dermatol 14:296–298 [DOI] [PubMed] [Google Scholar]
- Hemanthkumar, Thappa DM (1997) Dyschromatosis symmetrica hereditaria in an Indian family. J Dermatol 26:544–545 [DOI] [PubMed] [Google Scholar]
- Tomaya I (1929) Dyschromatosis symmetrica hereditaria (in Japanese). Jpn J Dermatol 29:95–96 [Google Scholar]
- Urabe K, Hori Y (1997) Dyschromatosis. Semin Cutan Med Surg 16:81–85 [DOI] [PubMed] [Google Scholar]
- Voron DA, Hatfield HH, Kalkhoff RK (1976) Multiple lentigines syndrome: case report and review of the literature. Am J Med 60:447–456 [DOI] [PubMed] [Google Scholar]
- Yang X, She C, Guo J, Yu AC, Lu Y, Shi X, Feng G, He L (2000) A locus for brachydactyly type A-1 maps to chromosome 2q35-q36. Am J Hum Genet 66:892–903 [DOI] [PMC free article] [PubMed] [Google Scholar]