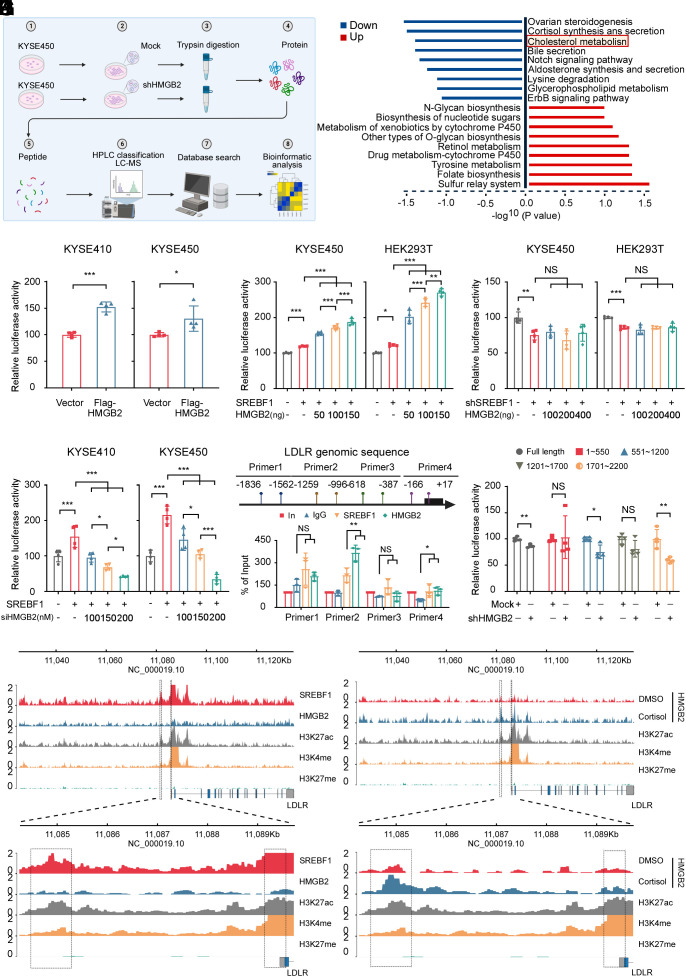
Fig. 4.
HMGB2 promotes ESCC proliferation through the cholesterol metabolism pathway. (A) Schematic illustrating the flow of the proteomic sequencing experiments using mock and shHMGB2 in KYSE450 cells. The figure was created with BioRender.com. (B) Differentially enriched protein list detected from the proteomic sequencing of shHMGB2 vs Mock KYSE450 cells. (C) Relative LDLR luciferase promoter activity in KYSE410 and KYSE450 cells overexpressing of HMGB2. Dual-luciferase was detected at 48 h after transfection. (n = 4/group). (D and E) Dual-luciferase reporter assay was used to measure the relative LDLR luciferase promoter activity in KYSE450 and HEK293T cells overexpressing (D) or depleted of SREBF1 (E) 48 h after cotransfecting the LDLR promoter-reporter plasmid and different concentrations of HMGB2. (n = 4/group). (F) Dual-luciferase reporter assay was used to measure the relative LDLR luciferase promoter activity in KYSE410 and KYSE450 cells overexpressing SREBF1 after cotransfecting the LDLR promoter-reporter and siHMGB2 plasmid 48 h. (n = 4/group). (G) Schematic illustrating the designed primers used to assess the enrichment of SREBF1 and HMGB2 at the LDLR promoter. The figure was created with BioRender.com. (H) The gray value analysis of SREBF1 and HMGB2 enrichment at the LDLR promoter was measured in KYSE450 cells using the CHIP assay. (n = 3/group). (I) Dual-luciferase reporter assays of KYSE450 cells transfected with LDLR truncated promoters or full length. (n = 4/group). (J) Gene tracks illustrating SREBF1, HMGB2, H3K27ac, H3K4me, and H3K27me enrichment at the LDLR promoter locus in KYSE450 cells based on CUT&TAG analysis. (K) Gene tracks illustrating HMGB2, H3K27ac, H3K4me, and H3K27me enrichment at the LDLR promoter locus in KYSE450 cells based on CUT&TAG analysis after treatment with DMSO or 10 μM cortisol for 18 h. *P < 0.05, **P < 0.01, ***P < 0.001, NS = no significance.