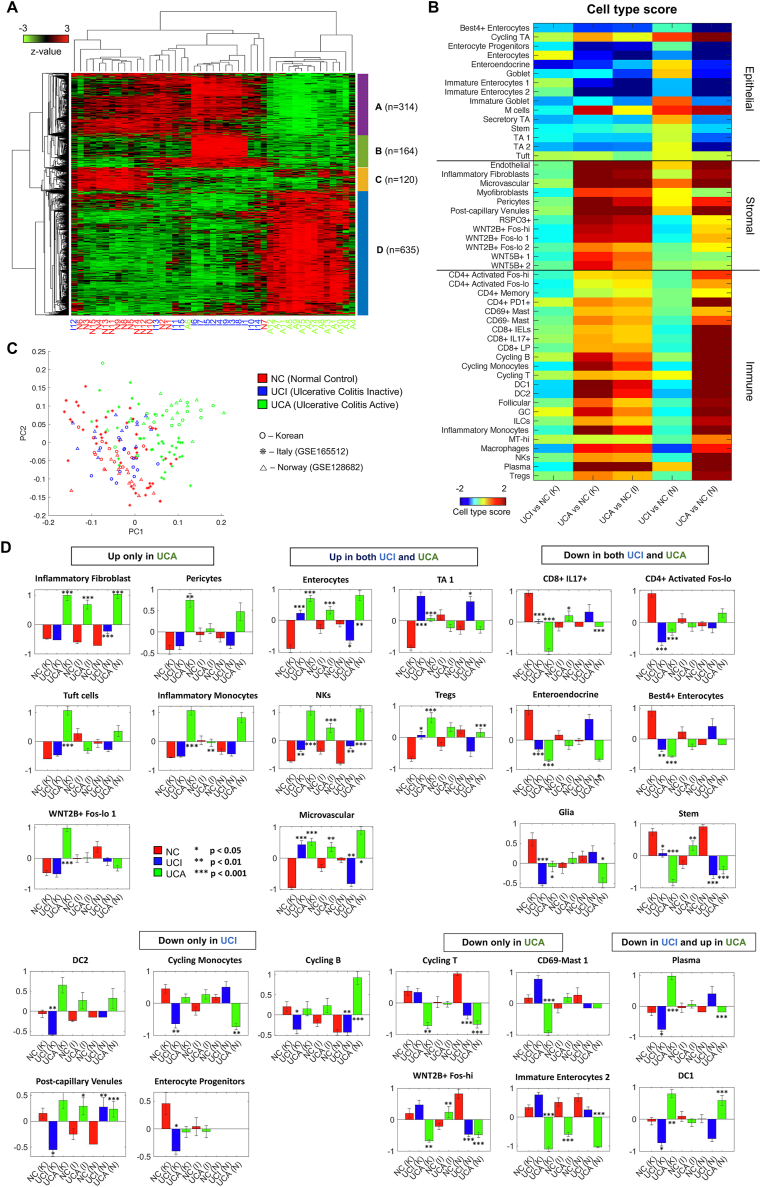
Figure 1.
Integrative analysis using bulk and single-cell RNA sequencing revealed inflammatory transcriptomic signatures in ulcerative colitis patients. (A) The heatmap of the differentially expressed genes sorted by two-way hierarchical clustering. The differentially expressed genes were clustered into four groups. (B) The principal component analysis plot of the three datasets from Korean (our study), Italian, and Norwegian studies. (C) A heatmap of the 50 cell type scores for each comparison of bulk RNA sequencing data of the three datasets. The cell type scores based on single-cell RNA sequencing suggest the putative cell type contributions in the bulk RNA sequencing data. (D) A total of 27 cell types were significantly altered in different patient types of the three datasets. Deconvolution of RNA sequencing reveals altered cell types in active inflammatory colonic tissue (UCA). The K, I, and N in the parenthesis represent Korean, Italian, and Norwegian datasets, respectively. UCI, macroscopically inactive inflammatory colonic tissue; NC, normal control.