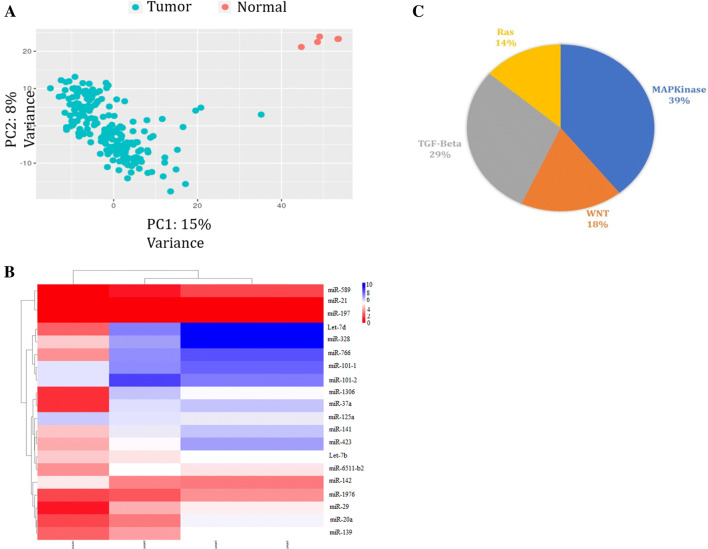
Fig. 1.
Selection of candidate CRC-related miRNAs through bioinformatics analysis of TCGA database. a PCA analysis showed correct differentiation of normal data from tumor data
adopted from TCGA database. b Drawn heat map for the top 20 miRNAs which are differentially expressed in normal and tumor CRC samples using heatmap2 package in R programing. c Target predictions of 10 candidate miRNAs. The MAPK and TGFβ signaling were the major pathways to be targeted by candidate miRNAs