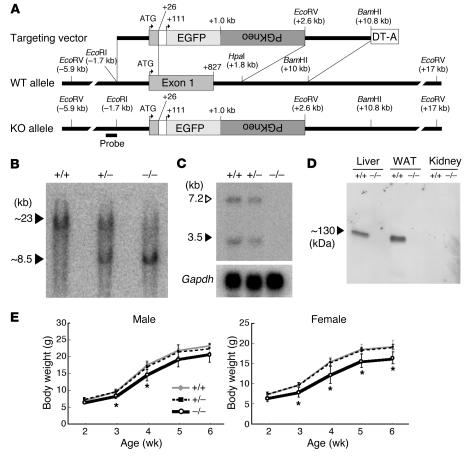
Figure 1.
Targeted disruption of the βklotho gene. (A) Strategy for disruption of mouse βklotho locus. Schematic diagrams of the targeting vector (top), wild-type allele (middle), and disrupted allele (bottom) are shown. Numbers correspond to the distance (bp) from the putative translation initiation site. The probe used for Southern blot analysis is shown at the bottom. (B) Southern blot analysis of EcoRV-digested genomic DNA. Blots were hybridized with the probe indicated in A. (C) Northern blot analysis of βklotho gene expression in the liver. The βklotho-specific probe hybridizes with 2 species of transcripts (1). (D) Western blot analysis of βKlotho protein expression. Total proteins (15 μg) extracted from indicated tissues were blotted on a PVDF membrane and incubated with a βKlotho-specific monoclonal antibody. βKlotho protein was detected only in βklotho-expressing tissues of wild-type mice and was absent in tissues of βklotho–/– mice. WAT, white adipose tissue. (E) Growth curve comparison of βklotho+/+, βklotho+/–, and βklotho–/– mice. Body weights were traced in mice from ages 2 to 6 weeks. Five to 12 mice for each group were examined. Error bars indicate SD. *P < 0.05.