Abstract
Ehlers-Danlos VIII (EDS-VIII) is an autosomal dominant disorder characterized by severe early-onset periodontal disease in conjunction with the features of Ehlers-Danlos syndrome (EDS). We performed a genomewide linkage search in a large Swedish pedigree with EDS-VIII and established linkage to a 7-cM interval on chromosome 12p13, generating a maximum multipoint LOD score of 5.17. Analysis of four further pedigrees with EDS-VIII revealed two consistent with linkage to 12p13 and two in which linkage could be excluded, indicating that EDS-VIII is a genetically heterogeneous disorder. Chromosome 12p13 has not previously been implicated in either EDS or periodontal disease and contains no known collagen genes or collagen-processing enzymes. Mutational screening of the microfibril-associated glycoprotein-2 gene, a strong candidate within the minimal interval, did not reveal any likely pathogenic mutations.
Ehlers-Danlos syndrome (EDS) is a clinically and genetically heterogeneous connective-tissue disorder characterized by articular hypermobility, skin hyperextensibility, and tissue fragility (McKusick 1972; Pope and Burrows 1997). Pathogenic mutations in the genes encoding collagen types I, III, and V, and the collagen-processing enzymes lysyl hydroxylase and procollagen N-peptidase have been found to underlie several EDS variants (Beighton et al. 1998), which suggests that EDS is a disorder of fibrillar collagen metabolism (Byers 1995). More recently, mutations in Tenascin-X, a large extracellular-matrix protein of unknown function, were reported in patients with EDS, indicating that defects outside the classic collagen family have potential to cause EDS phenotypes (Schalkwijk et al. 2001).
Ehlers-Danlos VIII (EDS-VIII [MIM 130080]) is an autosomal dominant condition in which severe premature periodontal disease segregates with otherwise typical EDS (Pope and Burrows 1997). Since the first report in 1967, <40 cases have been published (Barabas and Barabas 1967; Stewart et al. 1977; Linch and Acton 1979; Piette and Douniau 1980; Lapiere and Nusgens 1981; Nelson and King 1981; Olesen and Ernst 1987; Riedl et al. 1989; Flachowsky et al. 1990; Hartsfield and Kousseff 1990; Biesecker et al. 1991; Hoffman et al. 1991; Bond 1993; Cuniff and Williamson-Kruse 1995; Spranger et al. 1996; Karrer et al. 2000). The clinical manifestations in these patients vary, with differing degrees of skin hyperextensibility, fragility, and scarring; minimal-to-moderate joint hypermobility (usually limited to the digits); and normal or slightly increased tendency to bruising on mild trauma. Discrete, chronically inflamed pretibial plaques, reminiscent of necrobiosis lipoidica, are often present. The underlying molecular cause of EDS-VIII is unknown. A reduction of collagen type III was reported in a single case (Lapiere and Nusgens 1981), but no consistent biochemical or structural changes are detectable (Hartsfield and Kouseff 1990; Dyne et al. 1993).
The primary feature that discriminates EDS-VIII from other forms of EDS is severe early-onset periodontitis. Periodontitis is characterized by irreversible destruction of the periodontal tissues (periodontal ligament, alveolar bone, and connective tissue) and has many heterogeneous causes. It is estimated to occur in 15%–20% of the adult population and 0.5%–3% of children, depending on the population studied (Papapanou 1996). Most forms of periodontitis are thought to result from complex interactions of pathogenic genetic variants with environmental agents. However, specific single-gene defects can predispose to some forms of periodontal disease, particularly early-onset periodontitis, which is associated with a number of genetic syndromes, including chronic familial neutropenia (MIM 162700), leucocyte adhesion deficiency type II (MIM 266265), Chediak-Higashi syndrome (MIM 214500), and Papillon-Lefevre syndrome (MIM 245000).
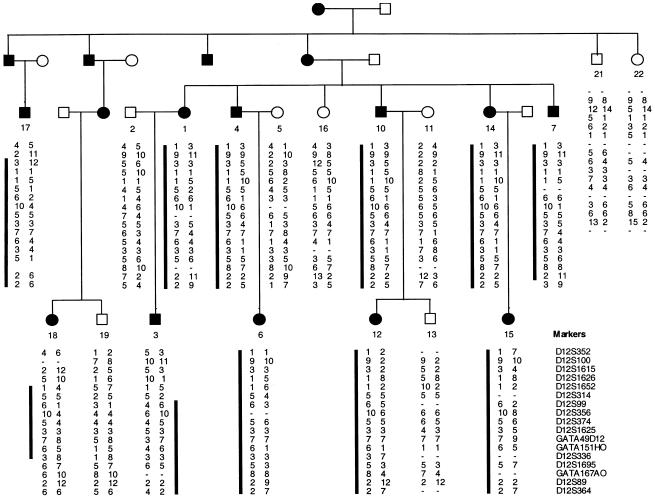
We ascertained a large five-generation family with EDS-VIII from Pitea in northwestern Sweden (fig. 1). This family is referred to as “family A.” Seventy-two individuals from five generations of family A were examined after full informed consent was obtained. Dermatological features were assessed by one of the authors (F.M.P.), and individuals were classified as affected using published clinical criteria for the diagnosis of EDS (Beighton et al. 1998). Independently, dental features were examined by another author (B.F.), and standard dental criteria were used to diagnose early-onset periodontitis. The affection status as determined by dermatological and dental criteria were entirely congruent in all individuals.
Figure 1.
Haplotypes for 17 markers from chromosome 12p13 in family A. Blackened symbol = affected with EDS-VIII. Unblackened symbol = unaffected individual. The black bar next to marker alleles indicates that the disease-segregating haplotype is present in affected individuals. Meiotic recombinants in individuals 3 and 18 define the minimal interval encompassing the EDS-VIII gene to the region between markers D12S314 and D12S1695.
The predominant dental features were premature periodontal inflammation and gingival recession, usually detectable in childhood, with rapid progression throughout adolescence and early adulthood. In general, complete loss of adult dentition occurred, typically, in individuals born prior to 1950, by the end of the third decade. Dental X-rays of selected family members showed progressive loss of periodontal supporting tissue and loss of bony tissue.
Affected individuals showed typical features of EDS, with generalized joint laxity (Beighton scores 5/9–9/9), thin atrophic skin, (especially over the dorsum of the hands and feet), and circumscribed hemosiderotic pretibial plaques, which could be thickened or atrophic. Affected individuals were tall, with spans wider than their heights.
Collagen protein analysis and transmission electron microscopy showed no evidence of collagen III deficiency. However, subtle irregularities in the pattern of dermal collagen fibrils, with an abnormally mixed pattern of larger and smaller fibrils, were observed (data not shown).
A genomewide linkage search was performed, using 11 affected individuals from family A. Four hundred microsatellite markers spaced at ∼10-cM intervals from ABI PRISM linkage-mapping set version 2 (Applied Biosystems) were PCR amplified, by use of standard protocols. Amplified markers were electrophoresed on an ABI 3770 DNA capillary sequencer and were analyzed with GENESCAN and GENOTYPER software (Applied Biosystems).
EDS-VIII was modeled as an autosomal dominant trait that is fully penetrant by age 15 years. Unaffected individuals under age 15 years were coded as “unknown.” A disease allele frequency of .001 and equal recombination fractions in males and females were assumed. Two-point LOD scores were calculated by use of the MLINK program of LINKAGE (Lathrop et al. 1984), and multipoint LOD scores (four markers and the disease) were generated by use of VITESSE (O’Connell and Weeks 1995). Marker allele frequencies were estimated from individuals in the pedigree.
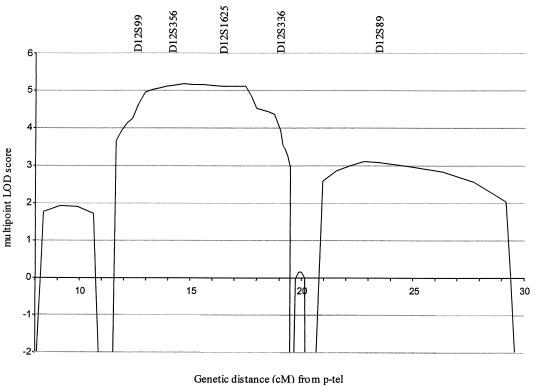
In the initial linkage search, marker D12S99 generated the highest two-point LOD score, and positive LOD scores at consecutive markers occurred only at D12S99 and D12S336 on chromosome 12p13. To confirm and refine the location of the EDS-VIII predisposition gene, 15 additional markers spanning a 30-cM interval on chromosome 12p were analyzed. Samples from five unaffected relatives and three spouses were included in these analyses (fig. 1). The order and distance between the markers were obtained from the Marshfield Genetic Database and are presented telomeric to centromeric, as follows: D12S352-3.3 cM-D12S100-1.3 cM-D12S1615-2.5 cM-D12S1626-0.5 cM-D12S1652-3.8 cM-D12S314-1.2 cM-D12S99-1.6 cM-D12S356-0 cM-D12S374-2.2 cM-D12S1625-1.3 cM-GATA49d12-0 cM-GATA151h0-1.3 cM-D12S336-0.6 cM-D12S1695-0.7 cM-GATA167a0-2.9 cM-D12S89-6.2 cM-D12S364. A haplotype of marker alleles segregates with the disease in all affected patients and is not present in any of the unaffected individuals (fig. 1). Critical meiotic recombinants in ID-3 (telomeric) and ID-18 (centromeric) place the EDS-VIII predisposition gene in a 7-cM region flanked by D12S314 and D12S1695 (fig. 1). The maximum two-point LOD score was 4.04 at D12S356 (table 1). The maximum multipoint LOD score was 5.17 close to D12S356 (fig. 2).
Table 1.
Two-Point LOD Scores at Chromosome-12p13 Markers in Swedish Pedigree with EDS-VIII
|
Two-point LOD Score at Θ |
||||||
| Marker | .00 | .05 | .1 | .2 | .3 | .4 |
| D12S352 | −∞ | −.24 | −.65 | −.80 | .61 | .27 |
| D12S100 | −∞ | −.08 | .29 | .43 | .32 | .11 |
| D12S1615 | −∞ | 1.88 | 1.86 | 1.45 | .89 | .32 |
| D12S1626 | −∞ | .84 | .95 | .80 | .47 | .13 |
| D12S1652 | −∞ | 3.34 | 3.69 | 2.88 | 1.96 | .87 |
| D12S314 | −∞ | .29 | .47 | .47 | .30 | .09 |
| D12S99 | 2.08 | 1.80 | 1.52 | .98 | .49 | .13 |
| D12S356 | 4.04 | 3.66 | 3.26 | 2.40 | 1.50 | .60 |
| D12S374 | 1.76 | 1.52 | 1.28 | .81 | .38 | .08 |
| D12S1625 | 1.39 | 1.30 | 1.20 | .96 | .69 | .37 |
| GATA49d12 | 1.39 | 1.25 | 1.09 | .76 | .44 | .17 |
| GATA151h0 | 2.76 | 2.53 | 2.27 | 1.66 | .99 | .38 |
| D12S336 | .711 | .60 | .49 | .28 | .12 | .02 |
| D12S1695 | −∞ | 1.10 | 1.12 | .85 | .45 | .13 |
| GATA167a0 | −∞ | 1.40 | 1.51 | 1.33 | .93 | .43 |
| D12S89 | 1.99 | 1.85 | 1.69 | 1.31 | .87 | .36 |
| D12S364 | −∞ | .64 | .95 | 0.94 | .62 | .21 |
Figure 2.
Multipoint LOD score at chromosome 12p13 in family A
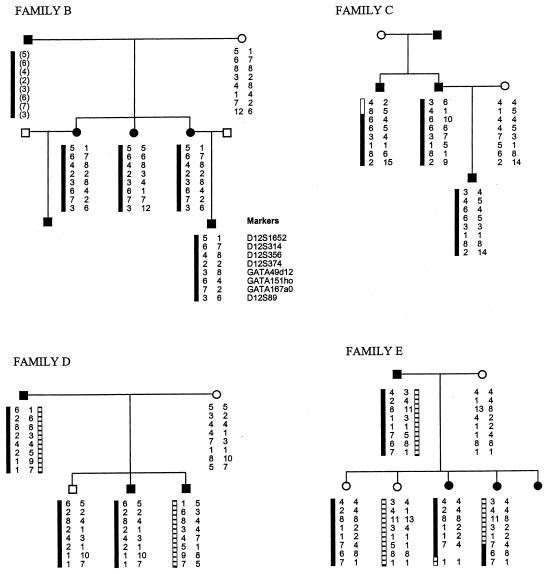
We analyzed a further four pedigrees with EDS-VIII (families B, C, D, and E) for linkage to chromosome 12p13 (fig. 3). Affected individuals from all four pedigrees were examined by one of the authors (F.M.P.) and were considered typical of EDS-VIII. Eight chromosome-12p13 microsatellite markers spanning the critical EDS-VIII interval were analyzed in each pedigree. In families D and E, the affected individuals do not share a chromosome-12p13 haplotype, and some unaffected individuals carry the same chromosome-12p13 haplotype as affected individuals (fig. 3). This excludes the EDS-VIII gene we have identified as the causative gene in these two families and demonstrates that EDS-VIII is a genetically heterogeneous disorder. Family B contains six affected individuals in three generations. DNA was available from four affected individuals, and all share a chromosome-12p13 haplotype that generates a multipoint LOD score of 0.42 at D12S356 (fig. 3). Similarly, family C contains three affected individuals and is also consistent with linkage to chromosome 12p13 generating a multipoint LOD score of 0.28 at D12S356. Thus, the occurrence of EDS-VIII in these two families may well be due to the chromosome 12p13 EDS-VIII gene, but the families are too small to give unequivocal evidence in favor of linkage in the context of known genetic heterogeneity.
Figure 3.
Haplotypes for eight chromosome-12p13 markers in four additional pedigrees with EDS-VIII (families B, C, D, and E). The segregating 12p13 haplotypes in affected individuals are depicted by black and hatched bars next to marker alleles. Inferred haplotypes are in parentheses.
Localization of an EDS-VIII gene to chromosome 12p13 has important implications. What is most important is that it provides the first clear evidence for the existence of EDS-VIII as a separate clinical entity. This has long been a point of contention, with some believing EDS-VIII to be a phenotypic variant of EDS-IV (MIM 130050), which is due to mutations in the COL3A1 gene on chromosome 2q31-32 (Hartsfield and Kouseff 1990; Dyne et al. 1993). Our data now provide molecular evidence that discriminates EDS-VIII from EDS-IV. This distinction has clinical implications for patients diagnosed with EDS-VIII, as it suggests they are unlikely to be at the same risk of the severe vascular complications seen in EDS-IV (Pepin et al. 2000).
Neither of the families unlinked to chromosome 12p13 has clinical features characteristic of EDS-IV. Furthermore, collagen protein analysis in family E demonstrated normal collagen III production and excretion, which precludes EDS-IV. Collagen analysis was not performed in family D. However, when microsatellite markers D2S103, D2S118, and D2S389, which closely flank COL3A1, were analyzed, they revealed that the affected sons have inherited opposite chromosome-2 haplotypes in this region from their affected father (data not shown). These data strongly suggest that COL3A1 mutations are not responsible for the EDS-VIII phenotype seen families D and E. In turn, this suggests that at least one further, currently unknown, EDS-VIII predisposition gene is likely to exist.
No other EDS-susceptibility gene has been mapped to chromosome 12p13, which indicates that a previously unrecognized gene can predispose to the Ehlers-Danlos phenotype. The genomic sequence of the critical interval is incomplete, but scrutiny of the UCSC Human Genome Project Working Draft indicates that the minimal interval is 4.6 Mb and contains 44 known genes and 16 anonymous and/or predicted genes. Within the minimal interval, there are no known collagen genes or collagen-modifying enzymes. The most promising candidate gene is microfibril-associated glycoprotein-2 (MAGP2) (Gibson et al. 1996). MAGP1 and MAGP2 are small, structurally related glycoproteins that specifically associate with fibrillin-containing microfibrils. Fibrillin-containing microfibrils are important structural components of the extracellular matrix of most connective tissues. Morphologically identical microfibrils are found in association with elastin in tissues such as the aorta and elastic ligaments and as elastin-free bundles in other tissues, including the periodontal ligament and skin. MAGP1 and MAGP2 each contain a characteristic cysteine-rich core motif that is thought to be involved, in interactions of MAGPs, with other components of the microfibril. In addition, the N-terminal region of MAGP2 contains an RGD motif and interacts with a wide range of cell types in an RGD-dependent manner via αVβ3 integrin (Gibson et al. 1999). Of interest, Tenascin-X also contains an RGD sequence and mediates interactions via the same integrin subunit combination (Elefteriou et al. 1999).
We screened genomic DNA from affected and unaffected individuals from families A, B, and C for mutations in MAGP2, by use of conformation sensitive gel electrophoresis (Ganguly et al. 1993). Primers were designed to amplify the 10 exons and intron-exon boundaries of MAGP2, by use of primer3 software (table 2). No likely pathogenic alterations were detected, and no sequence variants segregating with the EDS-VIII phenotype were identified in any family (data not shown). This suggests that MAGP2 is not an EDS-VIII–predisposition gene, although it is possible that we have failed to detect deleterious MAGP2 alterations, either owing to lack of sensitivity of the screening technique or because they result in alterations that are not detectable by our methods, such as genomic rearrangements or regulatory mutations.
Table 2.
MAGP2 Mutation Screening Primers and Size of PCR Product
| Exon | Forward Primer | Reverse Primer | Size(bp) |
| 1 | gcaactgcaaattcccatct | cattccctctgtctctttaggg | 284 |
| 2 | tgcctctgagcatcacattc | tcttcgacctcattccatcc | 193 |
| 3 | ccatttctccatgattttctga | tggcagcgatagataagcag | 228 |
| 4 | tggcagggagacagagaatc | cctccaacccttctgaagtc | 198 |
| 5 | tccccttatcctgctttcct | gggcaacagaatgagactcc | 259 |
| 6 | gccttgtgtttgcaatttga | tcagcagctgcacactatcc | 186 |
| 7 | gcactccaagcaggtcctaa | gtgccaagtaccctccaaag | 212 |
| 8 | cccaattccttcaatccttc | gcaattccatagtgggtgct | 251 |
| 9 | atcccagacccaggtgagtt | gcctgaagccctcctctagt | 259 |
| 10 | tctgcattctcttcccatga | cctccttcctctcacccata | 202 |
Mapping of the EDS-VIII gene also has implications for other forms of periodontal disease. Our data demonstrate that a gene predisposing to early-onset periodontal disease is located at chromosome 12p13. No genes within this chromosomal interval have previously been implicated in periodontal disease. Because the cutaneous features in pedigrees with EDS-VIII can be variable and sometimes subtle, it would be instructive to examine pedigrees with familial periodontal disease for linkage to chromosome 12p13. Isolation of the EDS-VIII gene will allow more focused investigations of its functions and is likely to greatly further our understanding of the molecular mechanisms that lead to both EDS and periodontal disease.
Acknowledgments
We are grateful to all the family members for their kind cooperation in this research. We thank Arne Zinkmark, for making clinical facilities available for family assessment; Gun-Britt Lundstrom, for assisting with ascertainment of blood samples; the members of the Cancer Genome Project who contributed to the genomewide search; and the Wellcome Trust. This research was supported by the Medical Research Council, the Ehlers-Danlos Support Group, and the Institute of Cancer Research.
Electronic-Database Information
URLs for data presented herein are as follows:
- Marshfield Genetic Database http://research.marshfieldclinic.org/genetics/ (for identification and order of microsatellite markers)
- Online Mendelian Inheritance in Man (OMIM) http://www.ncbi.nlm.nih.gov/Omim/ (for Ehlers-Danlos VIII, Ehlers-Danlos IV, chronic familial neutropenia, leucocyte adhesion deficiency type II, Chediak-Higashi syndrome, and Papillon-Lefevre syndrome)
- Primer3 http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi (for design of MAGP2 screening primers)
- UCSC Human Genome Project Working Draft, http://genome.cse.ucsc.edu/ (for identification of candidate genes)
References
- Barabas GM, Barabas AP (1967) The Ehlers-Danlos syndrome: a report of oral and haematological findings in nine cases. Br Dent J 123:473–479 [PubMed] [Google Scholar]
- Beighton P, De Paepe A, Steinmann B, Tsipouras P, Wenstrup RJ (1998) Ehlers-Danlos syndromes: revised nosology. Am J Med Genet 77:31–37 [DOI] [PubMed] [Google Scholar]
- Biesecker LG, Erickson RP, Glover TW, Bonadio J (1991) Molecular and cytologic studies of Ehlers-Danlos syndrome type VIII. Am J Med Genet 41:284–288 [DOI] [PubMed] [Google Scholar]
- Bond PJ, Friend GW, Meridith MW (1993) Ehlers-Danlos syndrome identified from periodontal findings: case report. Pediatr Dent 15:212–213 [PubMed] [Google Scholar]
- Byers P (1995) Disorders of collagen biosynthesis and structure. In: Scriver C, Beauder A Sly W, Valle D (eds) The metabolic basis of inherited disease. McGraw-Hill, New York, pp 4029–4075 [Google Scholar]
- Cunniff C, Williamson-Kruse (1995) Ehlers-Danlos syndrome type VIII presenting with periodontitis and prolonged bleeding time. Clin Dysmorphol 4:145–149 [PubMed] [Google Scholar]
- Dyne KM, Vitellaro-Zuccarello L, Bacchella L, Lanzi G, Cetta G (1993) Ehlers-Danlos type VIII: biochemical, stereological and immunocytochemical studies on dermis from a child with clinical signs of Ehlers-Danlos syndrome and a family history of premature loss of permanent teeth. Br J Dermatol 128:458–463 [DOI] [PubMed] [Google Scholar]
- Elefteriou F, Exposito JY, Garrone R, Lethias C (1999) Cell adhesion to tenascin-X mapping of cell adhesion sites and identification of integrin receptors. Eur J Biochem 263:840–848 [DOI] [PubMed] [Google Scholar]
- Flachowsky S, Tolkendorf E, Kamin G (1990) Ehlers-Danlos syndrom and Schwangenschaft. Zentralbl Gynakol 112:1369–1371 [PubMed] [Google Scholar]
- Ganguly A, Rock MJ, Prockop DJ (1993) Conformation-sensitive gel electrophoresis for rapid detection of single-base differences in double-stranded PCR products and DNA fragments: evidence for solvent-induced bends in DNA heteroduplexes. Proc Natl Acad Sci USA 90:10325–10329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson MA, Hatzinikolas G, Kumaratilake JS, Sandberg LB, Nichol JK, Sutherland GR, Cleary EG (1996) Further characterisation of proteins associated with elastic fiber microfibrils including the molecular cloning of MAGP-2. J Biol Chem 271:1096–1103 [DOI] [PubMed] [Google Scholar]
- Gibson MA, Leavesley DI, Ashman LK (1999) Microfibril-associated glycoprotein-2 specifically interacts with a range of bovine and human cell types via αVβ3 integrin. J Biol Chem 274:13060–13065 [DOI] [PubMed] [Google Scholar]
- Hartsfield JK Jr, Kousseff BG (1990) Phenotypic overlap of Ehlers-Danlos syndrome types IV and VIII. Am J Med Genet 37:465–470 [DOI] [PubMed] [Google Scholar]
- Hoffman GS, Filie JD, Schumacher HR Jr, Ortiz-Bravo E, Tsokos MG, Marini JC, Kerr GS, Ling QH, Trentham DE (1991) Intractable vasculitis, resorptive osteolysis, and immunity to type I collagen in type VIII Ehlers-Danlos syndrome. Arthritis Rheum 34:1466–1475 [DOI] [PubMed] [Google Scholar]
- Karrer S, Landthaler M, Schmalz G (2000) Ehlers-Danlos type VIII with severe periodontitis and apical root resorption after orthodontic treatment. Acta Derm Venereol 80:56–57 [DOI] [PubMed] [Google Scholar]
- Lapiere CM, Nusgens BV (1981) Ehlers-Danlos type VIII skin has a reduced proportion of collagen type III. J Invest Dermatol 76:422 [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J (1984) Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA 81:3443–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linch DC, Acton CH (1979) Ehlers-Danlos syndrome presenting with juvenile periodontitis. Br Dent J 147:95–96 [DOI] [PubMed] [Google Scholar]
- McKusick VA (1972) Heritable disorders of connective tissue, 4th ed. CV Mosby, St. Louis, pp 292–371 [Google Scholar]
- Nelson DL, King RA (1981) Ehlers-Danlos syndrome type VIII. J Am Acad Dermatol 5:297–303 [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE (1995) The VITESSE algorithm for rapid exact multilocus linkage analysis via genotype set-recoding and fuzzy inheritance. Nat Genet 11:402–408 [DOI] [PubMed] [Google Scholar]
- Olesen BH, Ernst E (1987) Orale manisfestioner ved Ehlers-Danlos syndrom type VIII. Tandlaegebladet 91:313–315 [PubMed] [Google Scholar]
- Papapanou PN (1996) Periodontal diseases: epidemiology. Ann Periodontol 1:1–36 [DOI] [PubMed] [Google Scholar]
- Pepin M, Schwarze U, Superti-Furga A, Byers PH (2000) Clinical and genetic features of Ehlers-Danlos syndrome type IV, the vascular type. N Engl J Med 342:673–680 [DOI] [PubMed] [Google Scholar]
- Piette E, Douniau R (1980) Parodontolyse infantile symptomatique d’un syndrome D’Ehlers-Danlos, un case sporadique? Acta Stom Belg 77:217–229 [PubMed] [Google Scholar]
- Pope FM, Burrows NP (1997) Ehlers-Danlos syndrome has varied molecular mechanisms. J Med Genet 34:400–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl M, Lost C, Pontz BF, Schofer O (1989) Ein Fall von Praubertarer Parodontitis: versuch einer Einordnung anhand von Laboruntersuchungen. Dtsch Zahnarztl Z 44:289–292 [PubMed] [Google Scholar]
- Schalkwijk J, Zweers MC, Steijlen PM, Dean WB, Taylor G, Van Vlijmen IM, Van Haren B, Miller WL, Bristow J (2001) A recessive form of the Ehlers-Danlos syndrome caused by tenascin-X deficiency. N Engl J Med 345:1167–1175 [DOI] [PubMed] [Google Scholar]
- Spranger S, Spranger M, Kirchof K, Steinmann B (1996) Ehlers-Danlos type VIII and leukodystrophy. Am J Med Genet 66:239–240 [DOI] [PubMed] [Google Scholar]
- Stewart RE, Hollister DW, Rimoin DL (1977) A new variant of Ehlers-Danlos syndrome: an autosomal dominant disorder of fragile skin, abnormal scarring, and generalized periodontitis. Birth Defects Orig Artic Ser 13:85–93 [PubMed] [Google Scholar]