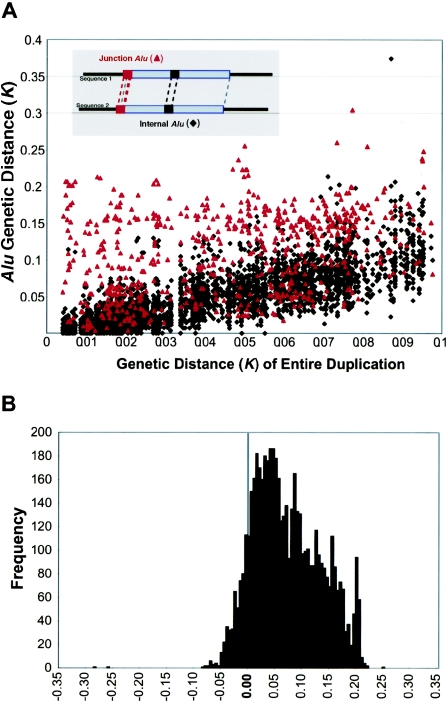
Figure 4.
Divergence of junction Alus. A, The sequence divergence of the segmental duplication (X-axis) is compared with the divergence of Alu repeats (Y-axis) for Alu repeats located internal to the pairwise alignment (black triangles) and Alu repeats localized at the junction (red triangles). Kimura’s two-parameter model of genetic distance (in changes/bp) is used as an estimate of divergence excluding CpG dinucleotides. Junction Alu repeats demonstrate an increased divergence relative to internal Alus. B, The pairwise differences in divergence between the junction Alus and each control Alu were calculated for each alignment (KjunctionAlu-KinternalAlu) (see the “Material and Methods” section). The alignment of the full-length Alu repeat element located at the junction, and not simply the Alu portion within the overall genomic alignment, was considered in this analysis. This measure shows a highly skewed positive distribution, with nearly 60% of all pairwise differences demonstrating a significant departure from that of an expected distribution (fig. B [online only]; >1 SD, based on a distribution of the difference between all possible combinations of internal Alu alignments).