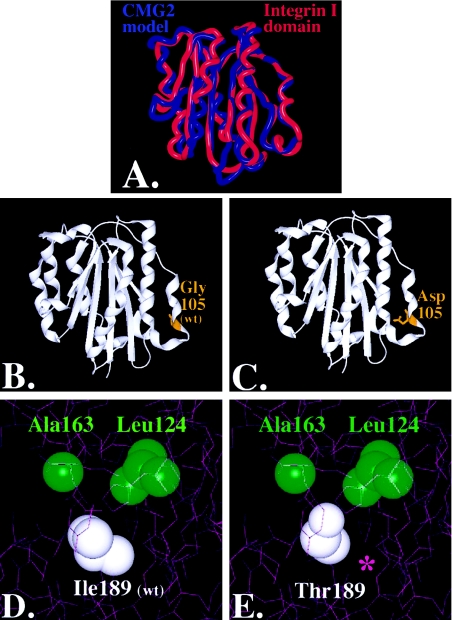
Figure 4.
Molecular modeling of CMG-2 mutations: superposition of CMG-2 model (red) with chain A of the Alpha-X Beta2 Integrin I Domain (PDB accession number 1N3Y) (blue). Nonconserved residues were mutated using the software program O (Jones et al. 1991), and the CMG-2 model was energy minimized using Molecular Operating Environment software (A). The root mean square deviation of the CMG-2 model from the integrin template is ∼.103 nm, with greater variation in the loops and less variance in the conserved regions where the mutations reside. Glycine 105 (B) was mutated to an aspartate (C), within the extracellular region and rendered with SPOCK and Raster3D (Merritt and Bacon 1997). Isoleucine 189 (D) was mutated to threonine (E), and contours were provided by the calculated electron density. A cavity is formed, as indicated by the purple asterisk (*) in panel E.