Abstract
To scrutinize the male ancestry of extant Native American populations, we examined eight biallelic and six microsatellite polymorphisms from the nonrecombining portion of the Y chromosome, in 438 individuals from 24 Native American populations (1 Na Dené and 23 South Amerinds) and in 404 Mongolians. One of the biallelic markers typed is a recently identified mutation (M242) characterizing a novel founder Native American haplogroup. The distribution, relatedness, and diversity of Y lineages in Native Americans indicate a differentiated male ancestry for populations from North and South America, strongly supporting a diverse demographic history for populations from these areas. These data are consistent with the occurrence of two major male migrations from southern/central Siberia to the Americas (with the second migration being restricted to North America) and a shared ancestry in central Asia for some of the initial migrants to Europe and the Americas. The microsatellite diversity and distribution of a Y lineage specific to South America (Q-M19) indicates that certain Amerind populations have been isolated since the initial colonization of the region, suggesting an early onset for tribalization of Native Americans. Age estimates based on Y-chromosome microsatellite diversity place the initial settlement of the American continent at ∼14,000 years ago, in relative agreement with the age of well-established archaeological evidence.
Introduction
Although there is general agreement that America was first settled from Asia by people who migrated across Beringia, the pattern of migration, its timing, and the place of origin in Asia of the people(s) that migrated to the Americas remain unclear (Fiedel 1992; Crawford 1998; Jablonski 2002). Synthesizing the linguistic, dental, and genetic information available at the time, Greenberg et al. (1986) proposed that the settlement of the American continent happened in three major migratory waves. According to this model, the first migration occurred ∼12,000 years ago (coinciding with the appearance of the Clovis cultural complex) and gave rise to the Amerind linguistic group, distributed across the Americas. The two subsequent migrations would be at the origin of two other major Native American language families: the Na Déne (in northwestern North America) and the Eskimo-Aleut (in circum-Arctic areas).
The three-migration-wave model of Greenberg et al. (1986) has since been taken as a working hypothesis for the interpretation of newer genetic data. In particular, the definition of molecular lineages through the use of arrays of linked polymorphisms now enables a phylogeographic analysis of human migrations into the Americas. This approach has been most extensively applied to mtDNA data and focuses on examining lineage distribution in populations from Asia and America, identifying founder lineages, and estimating their ages across linguistic groups and geographic regions. Four major founder Native American mtDNA haplogroups have been identified by RFLP and sequence analysis (Torroni et al. 1993; Forster et al. 1996; Merriwether and Ferrell 1996). The distribution and diversity of these haplogroups has usually been interpreted as indicative of more than one migratory wave during the initial colonization of the Americas (Torroni et al. 1992, 1993; Horai et al. 1993).
However, other mtDNA studies have challenged this interpretation and suggested the occurrence of a single migration into the continent, which would be at the origin of all major Native American linguistic families (Merriwether and Ferrell 1996; Bonatto and Salzano 1997a, 1997b; Silva et al. 2002). Interestingly, mtDNA analyses generally estimate an initial entry into the Americas prior to the last glacial maximum, ∼30,000 years before present (ybp) (Torroni et al. 1994; Forster et al. 1996; Bonatto and Salzano 1997a, 1997b; Silva et al. 2002). These age estimates have contributed to the debate generated by archaeological and geological findings concerning the possibility of a colonization of the Americas significantly earlier than assumed in Greenberg’s model (Crawford 1998; Jablonski 2002).
In the past few years, markers on the nonrecombining region of the Y chromosome have been used as a male complement to mtDNA analyses for the study of the colonization of the Americas. Initial analyses found one haplotype at high frequencies in native populations of all linguistic groups from North to South America (Pena et al. 1995; Santos et al. 1996; Underhill et al. 1996). This observation was interpreted as being indicative of a single founder Native American Y lineage, consistent with the view of a unique migratory wave into the continent. The other Y haplotypes detected in extant Native Americans were ascribed to recent admixture with nonnatives (Underhill et al. 1996; Bianchi et al. 1997). This putative single founder Native American lineage is characterized by a C→T mutation at marker M3 within the P-M45 Y lineage (Karafet et al. 1997; Underhill et al. 2001).
Calling into question the proposal of a single founder Y lineage in the Americas, the analysis of microsatellite (STR) diversity in five populations from Colombia indicated that a fraction of those carrying the ancestral M3 allele are Native American in origin (Ruiz-Linares et al. 1999). Moreover, in Asia, the M3 mutation has been found only in extreme northeastern Siberia, raising the possibility that this lineage is, in fact, native to America, from which it could have back migrated into neighboring Siberia (Karafet et al. 1997; Lell et al. 1997). Genuine Native American Y founder haplogroups would thus correspond to P-M45 lineages ancestral to M3 and possibly to lineages other than P-M45. Indeed, Bergen et al. (1999) recently identified a mutation (RPS4Y711) restricted to eastern Asia and America and marking a Native American founder lineage outside P-M45.
More recently, Karafet et al. (1999) and Lell et al. (2002) challenged the proposal of a single male migratory wave to the Americas and suggested the occurrence of at least two major migrations from northern Asia into America. According to Lell et al. (2002), the first migration introduced P-M45 chromosomes from southern central Siberia into the Americas, extending southwards to South America. The second migratory wave is proposed to have originated in eastern Siberia and to have reached only North and Central America. This second migration was delineated on the basis of P-M45 lineages characterized by STR alleles differentiated from those introduced in the first migration and by chromosomes carrying the RPS4Y711 mutation. A problem with the proposal of Lell et al. (2002) is that M45 chromosomes are found in both Native Americans and Europeans. This led Tarazona-Santos and Santos (2002) to suggest that the observation of differentiated P-M45 lineages in North/Central America compared with South America could relate to variable levels of recent admixture rather than to differences in the ancient demographic history of these population groups.
Recently, Seielstad et al. (2003 [in this issue]) identified a mutation (M242) ancestral to M3 and marking a Native American founder haplotype within the P-M45 lineage. Here, we report an analysis of Y-chromosome diversity (including the novel M242 polymorphism) in Native Americans and in a diverse sample from Mongolia, an important candidate region for the Asian source of the initial colonizers of the Americas (Neel et al. 1994; Kolman et al. 1996; Merriwether et al. 1996). The Native American populations typed include a Na Dené sample and a large set of South Amerinds. These data provide strong support for the occurrence of two major male migrations from central Siberia into America and are consistent with the existence of ancient links between proto-Europeans and proto–Native Americans. A lineage (Q-M19) with a restricted geographic distribution in South America has a diversity similar to that of its ancestor lineage Q-M3, suggesting an isolation of some Amerind populations soon after the initial dispersal of settlers in South America. In contrast to mtDNA analyses, Y-microsatellite diversity is more consistent with a relatively late colonization of the Americas.
Subjects and Methods
Populations Examined
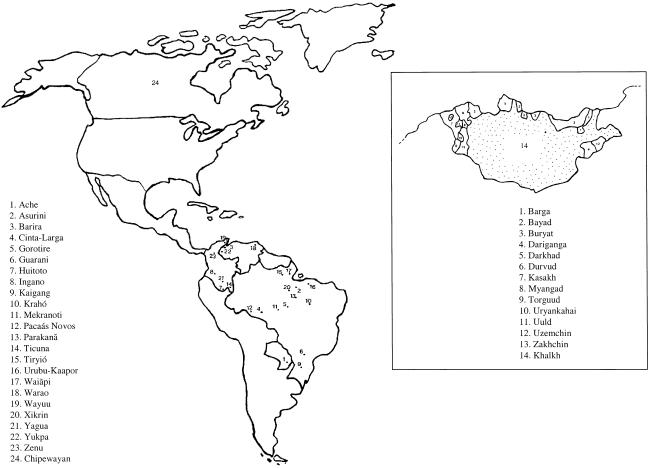
The total number of unrelated Native American males available for typing was 438 from 24 populations (figs. 1 and 2). The Na Dené linguistic family is represented by a broad sample of Chipewayan speakers (n=48) from several locations in Saskatchewan, Canada (primarily Fond-du-Lac and Uranium City). The South American populations were sampled in Brazil, Colombia, Paraguay, and Venezuela and include representatives of the four main subdivisions of South Amerind: Andean (Ingano, n=9), Chibchan-Paezan (Barira, n=12; and Warao, n=12), Equatorial Tucano (Ache, n=54; Asurini, n=4; Cinta-Larga, n=15; Guarani, n=59; Paacás Novos, n=15; Parakanã, n=20; Ticuna, n=33; Urubu-Kaapor, n=16; Waiãpi, n=14; and Wayuu, n=19), and Ge-Pano-Carib (Gorotire, n=5; Huitoto, n=4; Kaigang, n=22; Kraho, n=9; Mekranoti, n=7; Tiryió, n=4; Xikrin, n=8; Yagua, n=7; and Yukpa, n=12). The linguistic affiliation of one population (Zenu, n=30) is undefined (Mesa et al. 2000). The Mongolian sample (fig. 1) consisted mostly of Khalkh (n=303), Buryad (n=23), and Durvud (n=29), as well as a smaller number of individuals from 11 other ethnic groups (Barga, n=4; Bayad, n=10; Dariganga, n=1; Darkhad, n=4; Kazakh, n=6; Myangad, n=3; Torguud, n=3; Uryankhai, n=4; Uuld, n=2; Uzemchim, n=4; and Zakhchin, n=8). For most analyses, the 404 Mongolian individuals were considered as a single sample. Genomic DNA was extracted from whole blood, cheek cells, or plasma through use of the Qiagen Kit (QIAamp DNA Mini Kit), following the manufacturer’s instructions, or by organic phase extraction.
Figure 1.
Maps showing the approximate geographic location of the Native American populations examined (left) and the distribution of the ethnic groups sampled in Mongolia (inset).
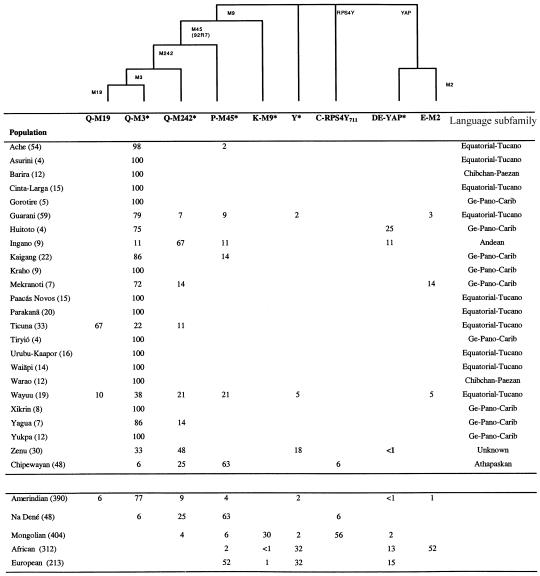
Figure 2.
Phylogenetic relationship of nine Y-chromosome lineages and their frequency in Native Americans, Mongolians, Europeans, and sub-Saharan Africans. The tree at the top displays the relationship of these lineages and the markers that were examined to identify them. The lineage frequency (%) in the populations examined here (with sample size in parentheses) is indicated in the middle. The mean lineage frequencies for major population groups are given at the bottom and were calculated from the data of Karafet et al. (1999) and Lell et al. (2002) and from the data collected here (the Na Dené being represented by the Chipewayan sample). The nomenclature of lineages follows the guidelines of the Y Chromosome Consortium (2002). Asterisks indicate lineages characterized by the derived state at that marker but the ancestral state at the other loci examined within that clade. For example, “P-M45*” refers to chromosomes carrying the derived M45 allele but the ancestral allele at marker M242—that is, P-M45*(x M242)—and Y* chromosomes carry the ancestral allele at all markers examined—that is, Y*(x M9, RPS4Y711, YAP).
Y-Chromosome Markers
Seven biallelic polymorphisms (M3 or DYS199, M19, 92R7, M9, YAP, M2 or DYS271, and RPS4Y711) were typed as described by Karafet et al. (1999), Ruiz-Linares et al. (1999), and Underhill et al. (2000). We also examined marker M242, recently identified by Seielstad et al. (2003 [in this issue]). Lineages defined by these mutations were designated following the recommendations of the Y Chromosome Consortium (2002). Six STR loci (DYS19, DYS388, DYS390, DYS391, DYS392, and DYS393) were genotyped fluorescently using the experimental conditions of Thomas et al. (1999). Data for these microsatellites in four of the Colombian populations had been reported elsewhere (Ruiz-Linares et al. 1999). Following common usage, we refer to microsatellite lineages as “haplotypes,” to distinguish them from biallelic marker “haplogroups.”
Data Analysis
Allele and lineage frequencies were calculated by counting, using the Arlequin package (Schneider et al. 2000). A principal-component analysis of Y-haplogroup frequencies was performed with the Ntsys (v. 2.1) program (Rohlf 2001). This analysis included data for the populations examined here, as well as data for the Asian and Native American populations examined by Karafet et al. (1999) and Lell et al. (2002). The relatedness of microsatellite haplotypes was assessed through reduced-median networks (Bandelt et al. 1995) constructed with the Network (v. 3.1) program. Neighbor-joining trees relating haplogroups on the basis of Nei’s genetic distance (Nei 1987), calculated from microsatellite allele frequencies, were obtained using programs in the Phylip (v. 3.5) package (Felsenstein 2001). The age of Y-chromosome haplogroups was estimated by calculating the mean average square distance (ASD) between the inferred ancestral haplotype of a haplogroup and all its observed descendants (Goldstein et al. 1995; Slatkin 1995; Ruiz-Linares et al. 1999). If we assume a generation time (g) of 25 years and a mean mutation rate (m) for Y microsatellites of 0.18% per generation (Kayser et al. 2000), the age of a haplogroup can be estimated as ASD×g/m. SEs for haplogroup age were obtained by bootstrapping the sample of observed haplotypes 1,000 times.
Results
Geographic Distribution of Y-Chromosome Haplogroups
The biallelic markers typed identified nine Y haplogroups in the populations examined (fig. 2). Four of these haplogroups (Q-M19, Q-M3*, Q-M242*, and C-RPS4Y711) are restricted to the Americas and/or Asia. The most derived haplogroup (Q-M19) was found only in the Ticuna and in the Wayuu of northwestern South America. Its parental haplogroup (Q-M3) was detected in all of the South Amerind populations tested, where it is markedly predominant (with a mean frequency of 77%). This haplogroup was also observed, albeit at a low frequency (6%), in the Chipewayan but was not found in Mongolia. Haplogroup Q-M242* (the immediate ancestor of Q-M3) is the second most prevalent in the Chipewayan, where it was observed at a frequency of 25%. This haplogroup has a wide distribution in South Amerinds (with a mean frequency of 9%), and its highest prevalence is in populations from northwestern South America (Ingano, Wayuu, and Zenu; figs. 1 and 2). Q-M242* was also detected at a low frequency (4%) in Mongolia. Haplogroup C-RPS4Y711 was not found in South Amerinds, has a low frequency (6%) in the Chipewayan, and is the lineage most commonly observed in Mongolia (56%).
The other five lineages detected (P-M45*, K-M9*, Y*, DE-YAP, and E-M2) are observed outside the Americas and Asia (fig. 2). Lineage P-M45* was found at a low frequency in South Amerinds (4%) and Mongolia (6%) but is the most common in Europe (52%) and the Chipewayan (63%). Lineage K-M9* has low frequencies in Europe and Africa, is the second most frequent in Mongolia (30%), and was not observed in Native Americans. Lineages Y*, DE-YAP, and E-M2 are mostly restricted to Europe or Africa. They were not observed in the Chipewayan, and their low frequency in South Amerinds (∼4%) is likely to reflect recent admixture.
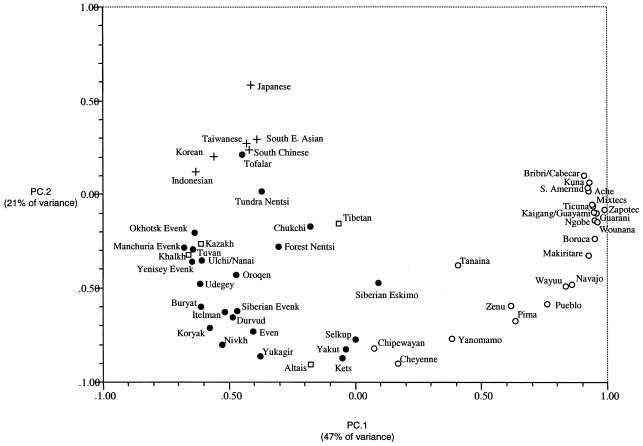
Figure 3 shows a principal-component analysis of the frequency of six Y-chromosome haplogroups in the populations typed here and in the Asian and Native American populations examined by Karafet et al. (1999) and Lell et al. (2002). The first principal component separates Native American from Asian populations, with the Chipewayan, the Cheyenne, and several populations from northern Asia occupying intermediate positions on this axis. The second principal component separates populations within America and Asia, with the Chipewayan, the Cheyenne, and some northern Asian populations (Selkup, Yakut, and Kets) maintaining a close genetic affinity. The spread of Native American populations on the principal-component graph correlates mostly with the frequency of haplogroups Q-M3 and P-M45* (data not shown), with populations from South America generally having the highest frequencies of Q-M3, whereas some populations from North America show increased frequencies of P-M45* and also show the presence of haplogroup C-RPS4Y711. Similarly, the Asian populations closest to Native Americans are characterized by a predominance of lineage P-M45* and low frequencies of C-RPS4Y711. Northeastern Asian populations generally appear closely related to central Asians, reflecting the generally high frequency of haplogroups K-M9* and C-RPS4Y711 in most of these populations (data not shown).
Figure 3.
Principal-component analysis based on the frequency of six Y-chromosome lineages (Q-M3, P-M45*, C-RPS4Y711, K-M9, DE-YAP, and Y*) in 64 Asian and Native American populations (with sample sizes >10). For the populations typed here, the frequency of haplogroups Q-M19 and Q-M242* shown in figure 2 was added to that of lineages Q-M3* and P-M45*, respectively. The Mongolian data collected here were analyzed separately for the Khalkh, Buryad, and Durvud samples. Data for the other populations are from Karafet et al. (1999) and Lell et al. (2002). Eastern Asians are shown as crosses, central Asians as squares, northern Asians as blackened circles, and Native Americans as unblackened circles. “S. Amerind” refers to nine South American populations in which the only lineage observed is Q-M3.
STR Diversity of Y-Chromosome Haplogroups
Haplogroups Q-M3* and Q-M19
A total of 36 Q-M3* STR haplotypes were identified in South Amerinds (table 1). Twenty-two of these haplotypes were seen only once. Of the 14 haplotypes seen multiple times, 9 were seen in only one population, and two or three populations share the other 5. The high frequency of haplotype 1 is mostly due to this being the only haplotype observed in the Ache (n=54), a population with a marked reduction in Y-chromosome diversity. A median-joining network relating Q-M3* haplotypes detected more than once is shown in figure 4A. At the center of this network is haplotype 5. This haplotype contains the most frequent allele at the six STR loci tested (table 2). The data of Lell et al. (2002) for four of the six STR loci examined here (DYS19, DYS388, DYS390, and DYS391) show that a haplotype characterized by alleles 13-12-24-10 (included in haplotype 5 in table 1) has a wide distribution in North and Central Amerinds. These observations confirm our previous results (Ruiz-Linares et al. 1999) identifying haplotype 5 as the most likely ancestor of Q-M3* chromosomes and lead to an estimated age for this haplogroup of 7,570 years (SE 681).
Table 1.
Q-M3* and Q-M19 Haplotypes in Native Americans
|
Allele at |
Frequency |
|||||||
| Haplotype | DYS19 | DYS388 | DYS390 | DYS391 | DYS392 | DYS393 | Q-M3*(n=117) | Q-M19(n=18) |
| 1 | 13 | 12 | 23 | 10 | 14 | 13 | .265 | .056 |
| 2 | 13 | 12 | 24 | 10 | 13 | 14 | .094 | 0 |
| 3 | 13 | 12 | 24 | 10 | 15 | 12 | .086 | 0 |
| 4 | 14 | 12 | 23 | 10 | 14 | 13 | .051 | 0 |
| 5 | 13 | 12 | 24 | 10 | 14 | 13 | .051 | 0 |
| 6 | 14 | 13 | 24 | 10 | 14 | 13 | .043 | 0 |
| 7 | 13 | 12 | 24 | 10 | 15 | 14 | .034 | 0 |
| 8 | 14 | 12 | 23 | 10 | 13 | 13 | .034 | 0 |
| 9 | 13 | 12 | 24 | 11 | 14 | 13 | .034 | 0 |
| 10 | 13 | 12 | 24 | 10 | 13 | 12 | .026 | 0 |
| 11 | 14 | 12 | 24 | 10 | 14 | 13 | .026 | 0 |
| 12 | 14 | 12 | 24 | 10 | 13 | 14 | .026 | 0 |
| 13 | 13 | 14 | 24 | 10 | 14 | 14 | .026 | 0 |
| 14 | 13 | 12 | 25 | 10 | 14 | 13 | .017 | .056 |
| 15 | 13 | 12 | 25 | 10 | 15 | 13 | .009 | .222 |
| 16 | 13 | 12 | 24 | 10 | 15 | 13 | .009 | .056 |
| 17 | 13 | 13 | 25 | 10 | 14 | 13 | .009 | .056 |
| 18 | 13 | 12 | 25 | 11 | 11 | 13 | .009 | 0 |
| 19 | 13 | 13 | 24 | 10 | 11 | 13 | .009 | 0 |
| 20 | 13 | 12 | 24 | 10 | 13 | 13 | .009 | 0 |
| 21 | 12 | 12 | 25 | 10 | 14 | 13 | .009 | 0 |
| 22 | 13 | 11 | 24 | 10 | 16 | 16 | .009 | 0 |
| 23 | 14 | 9 | 24 | 10 | 16 | 13 | .009 | 0 |
| 24 | 13 | 13 | 24 | 10 | 14 | 13 | .009 | 0 |
| 25 | 13 | 12 | 24 | 10 | 14 | 14 | .009 | 0 |
| 26 | 14 | 12 | 24 | 11 | 14 | 13 | .009 | 0 |
| 27 | 14 | 14 | 24 | 10 | 14 | 14 | .009 | 0 |
| 28 | 13 | 12 | 24 | 9 | 15 | 12 | .009 | 0 |
| 29 | 14 | 12 | 25 | 11 | 16 | 13 | .009 | 0 |
| 30 | 14 | 15 | 23 | 10 | 16 | 14 | .009 | 0 |
| 31 | 13 | 13 | 24 | 11 | 14 | 11 | .009 | 0 |
| 32 | 13 | 12 | 22 | 10 | 14 | 13 | .009 | 0 |
| 33 | 13 | 12 | 23 | 10 | 14 | 14 | .009 | 0 |
| 34 | 14 | 13 | 24 | 10 | 13 | 13 | .009 | 0 |
| 35 | 13 | 12 | 24 | 11 | 13 | 13 | .009 | 0 |
| 36 | 13 | 12 | 23 | 11 | 14 | 13 | .009 | 0 |
| 37 | 13 | 12 | 24 | 10 | 14 | 12 | 0 | .111 |
| 38 | 13 | 12 | 25 | 11 | 15 | 13 | 0 | .111 |
| 39 | 14 | 12 | 25 | 10 | 14 | 16 | 0 | .056 |
| 40 | 15 | 13 | 25 | 10 | 14 | 14 | 0 | .056 |
| 41 | 14 | 13 | 25 | 10 | 14 | 14 | 0 | .056 |
| 42 | 13 | 12 | 25 | 10 | 15 | 12 | 0 | .056 |
| 43 | 13 | 12 | 25 | 10 | 15 | 14 | 0 | .056 |
| 44 | 15 | 12 | 25 | 10 | 11 | 14 | 0 | .056 |
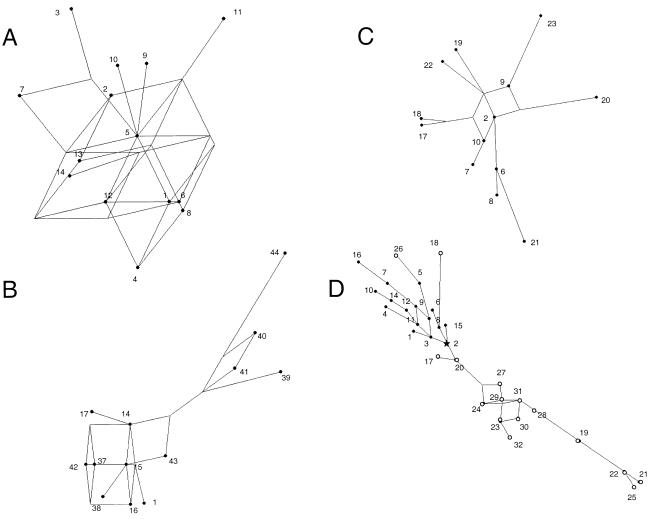
Figure 4.
Median-joining networks relating haplotypes within lineages Q-M3* (A), Q-M19 (B), Amerind Q-M242* (C), and P-M45* (D). Haplotype numbers are as indicated in tables 1, 3, and 4. In D, Native American haplotypes are shown as blackened circles, and Mongolian haplotypes are shown as unblackened circles. The star highlights a haplotype seen both in Native Americans and Mongolians.
Table 2.
Allele Frequencies at Six Microsatellite Loci in Lineages within the P-M45 Haplogroup in Amerindians, Chipewayan, and Mongolians
|
Allele Frequency |
||||||||
| Q-M242* |
P-M45* |
|||||||
| LocusandAllele | Q-M19 Amerindian(n=18) | Q-M3* Amerindian(n=117) | Amerindian(n=17) | Chipewayan(n=10) | Mongolian(n=15) | Amerindian(n=10) | Chipewayan(n=21) | Mongolian(n=23) |
| DYS19: | ||||||||
| 12 | 0 | .009 | .059 | 0 | 0 | 0 | 0 | 0 |
| 13 | .778 | .761 | .647 | .900 | .867 | .100 | .048 | 0 |
| 14 | .111 | .231 | .235 | 0 | .067 | .900 | .714 | .348 |
| 15 | .111 | 0 | .059 | .100 | .067 | 0 | .238 | .217 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | .261 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | .174 |
| DYS388: | ||||||||
| 9 | 0 | .009 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11 | 0 | .009 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | .833 | .855 | .765 | .900 | 1.000 | 1.000 | 1.000 | .913 |
| 13 | .167 | .086 | .177 | .100 | 0 | 0 | 0 | .087 |
| 14 | 0 | .034 | .059 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | .009 | 0 | 0 | 0 | 0 | 0 | 0 |
| DYS390: | ||||||||
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | .130 |
| 22 | 0 | .009 | 0 | .100 | .067 | .100 | .048 | .044 |
| 23 | .056 | .376 | .177 | .800 | .333 | .400 | .143 | .174 |
| 24 | .167 | .556 | .471 | .100 | .600 | .300 | .810 | .174 |
| 25 | .778 | .060 | .235 | 0 | 0 | .200 | 0 | .478 |
| 26 | 0 | 0 | .059 | 0 | 0 | 0 | 0 | 0 |
| 27 | 0 | 0 | .059 | 0 | 0 | 0 | 0 | 0 |
| DYS391: | ||||||||
| 5 | 0 | 0 | 0 | 0 | .067 | 0 | 0 | 0 |
| 6 | 0 | 0 | 0 | 0 | .133 | 0 | 0 | 0 |
| 9 | 0 | .009 | 0 | .100 | .200 | 0 | .048 | .044 |
| 10 | .889 | .906 | .882 | .300 | .467 | .400 | .095 | .565 |
| 11 | .111 | .086 | .118 | .600 | .067 | .600 | .857 | .391 |
| 12 | 0 | 0 | 0 | 0 | .067 | 0 | 0 | 0 |
| DYS392: | ||||||||
| 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | .174 |
| 11 | .056 | .017 | 0 | .100 | 0 | 0 | 0 | .478 |
| 12 | 0 | 0 | .059 | 0 | 0 | 0 | 0 | .044 |
| 13 | 0 | .205 | .118 | 0 | 0 | .600 | .571 | .304 |
| 14 | .444 | .598 | .588 | .700 | .800 | .300 | .429 | 0 |
| 15 | .500 | .145 | .177 | .200 | .067 | 0 | 0 | 0 |
| 16 | 0 | .034 | .059 | 0 | .067 | .100 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | .067 | 0 | 0 | 0 |
| DYS393: | ||||||||
| 11 | 0 | .009 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | .167 | .120 | .059 | .100 | 0 | 0 | 0 | .044 |
| 13 | .556 | .650 | .824 | .900 | .533 | .800 | .667 | .696 |
| 14 | .222 | .214 | .118 | 0 | .467 | .200 | .333 | .261 |
| 16 | .056 | .009 | 0 | 0 | 0 | 0 | 0 | 0 |
Table 1 also shows the 13 Q-M19 haplotypes observed in the Ticuna and the Wayuu. Haplotype 15 (seen four times and present in two Ticuna settlements) is the most frequent and is identical to an Q-M3* chromosome also found in the Ticuna. This haplotype includes the most frequent alleles at Q-M19 chromosomes (table 2) and is at the center of a network relating Q-M19 haplotypes (fig. 4B). Taking haplotype 15 as the ancestor of the Q-M19 lineage leads to an estimated age for this haplogroup of 7,972 years (SE 2,916).
Haplogroup Q-M242*
Twenty-five STR haplotypes were detected among Q-M242* chromosomes (table 3). One haplotype was shared between Mongolians and the Chipewayan and another between Amerindians and the Chipewayan, but none were shared between Amerindians and Mongolians. Ten of the Q-M242* haplotypes were also seen in lineages Q-M3* or Q-M19. Six of these Q-M242* haplotypes were found in Amerindians, four in the Chipewayan, and two in Mongolians. The modal haplotype in Amerindian Q-M242* chromosomes (haplotype 2) was found in three populations, contains the most common allele at each locus (table 2), and is at the center of a network relating haplotypes within this lineage (fig. 4C). Q-M242 haplotype 2 is also identical to the proposed ancestral haplotype of the Q-M3* lineage (haplotype 5 in table 1). A differentiation of lineage Q-M242* was observed among Amerindians, the Chipewayan, and Mongolians. The modal haplotype seen in Mongolians is one mutational step from the modal haplotypes of Amerindian and Chipewayan chromosomes, these last two being two mutational steps from each other (table 3). These haplotypes carry the most frequent alleles at each locus, and taking them as the most likely ancestors of Q-M242* chromosomes in these populations results in age estimates for this haplogroup in Mongolians, Amerinds, and the Chipewayan of 15,416 years (SE 4,722), 13,611 years (SE 2,916) and 6,014 years (SE 4,236), respectively.
Table 3.
Q-M242* Haplotypes in Mongolians, Chipewayan, and Amerindians
|
Allele at |
Frequency in |
||||||||
| Haplotype | DYS19 | DYS388 | DYS390 | DYS391 | DYS392 | DYS393 | Mongolian(n=15) | Chipewayan(n=10) | Amerindian(n=17) |
| 1 | 13 | 12 | 23 | 10 | 14 | 13 | .332 | .100 | 0 |
| 2 | 13 | 12 | 24 | 10 | 14 | 13 | 0 | .100 | .292 |
| 3 | 13 | 12 | 24 | 10 | 14 | 14 | .067 | 0 | 0 |
| 4 | 13 | 12 | 23 | 11 | 14 | 13 | 0 | .400 | 0 |
| 5 | 13 | 12 | 22 | 10 | 14 | 13 | 0 | .100 | 0 |
| 6 | 14 | 12 | 23 | 10 | 14 | 13 | 0 | 0 | .059 |
| 7 | 13 | 12 | 24 | 10 | 15 | 14 | 0 | 0 | .059 |
| 8 | 14 | 12 | 23 | 10 | 13 | 13 | 0 | 0 | .059 |
| 9 | 13 | 13 | 25 | 10 | 14 | 13 | 0 | 0 | .059 |
| 10 | 13 | 12 | 24 | 10 | 15 | 13 | 0 | 0 | .059 |
| 11 | 13 | 12 | 24 | 9 | 14 | 14 | .200 | 0 | 0 |
| 12 | 13 | 12 | 24 | 6 | 14 | 14 | .133 | 0 | 0 |
| 13 | 14 | 12 | 24 | 11 | 16 | 13 | .067 | 0 | 0 |
| 14 | 15 | 12 | 24 | 12 | 17 | 13 | .067 | 0 | 0 |
| 15 | 13 | 12 | 22 | 10 | 15 | 13 | .067 | 0 | 0 |
| 16 | 13 | 12 | 24 | 5 | 14 | 14 | .067 | 0 | 0 |
| 17 | 14 | 12 | 25 | 10 | 16 | 13 | 0 | 0 | .059 |
| 18 | 15 | 12 | 25 | 10 | 15 | 13 | 0 | 0 | .059 |
| 19 | 13 | 12 | 27 | 10 | 14 | 13 | 0 | 0 | .059 |
| 20 | 13 | 13 | 24 | 11 | 12 | 13 | 0 | 0 | .059 |
| 21 | 14 | 14 | 23 | 10 | 14 | 14 | 0 | 0 | .059 |
| 22 | 12 | 12 | 25 | 10 | 13 | 13 | 0 | 0 | .059 |
| 23 | 13 | 13 | 26 | 11 | 14 | 12 | 0 | 0 | .059 |
| 24 | 13 | 12 | 23 | 11 | 15 | 13 | 0 | .200 | 0 |
| 25 | 15 | 13 | 23 | 9 | 14 | 12 | 0 | .100 | 0 |
Haplogroup P-M45*
Table 4 shows the STR haplotypes detected in P-M45* chromosomes of Mongolians, the Chipewayan, and Amerinds. Haplotype 2 is shared across populations, with haplotype 3 (a one-step neighbor of haplotype 2) being shared between the Chipewayan and Amerinds. Mongolians show a markedly higher diversity of P-M45* haplotypes than do Native Americans, as indicated by a mean variance in repeat score of P-M45* chromosomes in Mongolia (1.22) that is three and six times higher than in Amerinds (0.41) and the Chipewayan (0.21), respectively (table 4). The higher diversity of Mongolian P-M45 haplotypes is also apparent in the network analysis in figure 4D, where some of the P-M45* haplotypes detected in Mongolia (2, 17, 18, 20, and 26) are found to be more closely related to haplotypes seen in Native Americans than to other Mongolian haplotypes.
Table 4.
P-M45* Haplotypes in Mongolians, Chipewayan, and Amerindians
|
Allele at |
Frequency in |
||||||||
| Haplotype | DYS19 | DYS388 | DYS390 | DYS391 | DYS392 | DYS393 | Mongolian(n=23) | Chipewayan(n=21) | Amerindian(n=10) |
| 1 | 14 | 12 | 25 | 11 | 13 | 13 | 0 | 0 | .200 |
| 2 | 14 | 12 | 24 | 10 | 13 | 13 | .044 | .048 | .100 |
| 3 | 14 | 12 | 24 | 11 | 13 | 13 | 0 | .286 | .100 |
| 4 | 14 | 12 | 24 | 11 | 16 | 13 | 0 | 0 | .100 |
| 5 | 14 | 12 | 22 | 11 | 13 | 14 | 0 | 0 | .100 |
| 6 | 14 | 12 | 23 | 10 | 14 | 13 | 0 | 0 | .200 |
| 7 | 13 | 12 | 23 | 11 | 14 | 14 | 0 | 0 | .100 |
| 8 | 14 | 12 | 23 | 10 | 13 | 13 | 0 | 0 | .100 |
| 9 | 14 | 12 | 24 | 11 | 14 | 14 | 0 | .190 | 0 |
| 10 | 15 | 12 | 23 | 11 | 13 | 13 | 0 | .095 | 0 |
| 11 | 14 | 12 | 24 | 11 | 14 | 13 | 0 | .048 | 0 |
| 12 | 15 | 12 | 24 | 11 | 14 | 13 | 0 | .095 | 0 |
| 13 | 14 | 12 | 24 | 11 | 13 | 14 | 0 | .095 | 0 |
| 14 | 15 | 12 | 23 | 11 | 14 | 13 | 0 | .048 | 0 |
| 15 | 14 | 12 | 24 | 9 | 13 | 13 | 0 | .048 | 0 |
| 16 | 13 | 12 | 22 | 10 | 14 | 14 | 0 | .048 | 0 |
| 17 | 15 | 12 | 24 | 10 | 13 | 12 | .044 | 0 | 0 |
| 18 | 14 | 12 | 19 | 10 | 13 | 13 | .130 | 0 | 0 |
| 19 | 16 | 12 | 23 | 11 | 10 | 14 | .044 | 0 | 0 |
| 20 | 15 | 12 | 24 | 10 | 13 | 13 | .044 | 0 | 0 |
| 21 | 14 | 13 | 23 | 10 | 10 | 14 | .044 | 0 | 0 |
| 22 | 14 | 12 | 23 | 10 | 10 | 14 | .044 | 0 | 0 |
| 23 | 17 | 12 | 25 | 10 | 11 | 13 | .044 | 0 | 0 |
| 24 | 15 | 12 | 25 | 10 | 11 | 13 | .087 | 0 | 0 |
| 25 | 14 | 12 | 23 | 9 | 10 | 14 | .044 | 0 | 0 |
| 26 | 14 | 13 | 22 | 11 | 13 | 13 | .044 | 0 | 0 |
| 27 | 16 | 12 | 24 | 10 | 11 | 13 | .044 | 0 | 0 |
| 28 | 16 | 12 | 25 | 11 | 11 | 14 | .087 | 0 | 0 |
| 29 | 15 | 12 | 25 | 11 | 11 | 13 | .044 | 0 | 0 |
| 30 | 17 | 12 | 25 | 11 | 11 | 13 | .087 | 0 | 0 |
| 31 | 16 | 12 | 25 | 11 | 11 | 13 | .087 | 0 | 0 |
| 32 | 17 | 12 | 25 | 10 | 12 | 13 | .044 | 0 | 0 |
| Variancea | 1.218 | .213 | .409 | ||||||
Mean variance in repeat score.
Relatedness of P-M45 Chromosomes across Populations
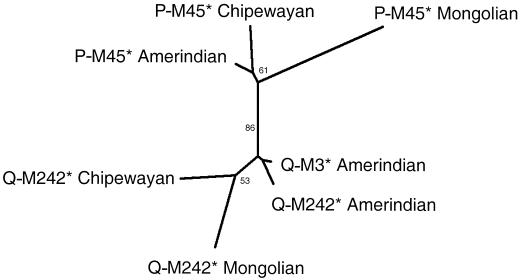
Figure 5 shows a neighbor-joining tree relating Q-M3*, Q-M242*, and P-M45* chromosomes in Amerinds, Chipewayan, and Mongolians, on the basis of genetic distances calculated from the allele frequencies observed at the six microsatellite loci tested (table 2). A clear separation is seen between chromosomes carrying the ancestral and derived alleles at marker M242. Haplogroup Q-M3* is found to be closely related to Amerind Q-M242* chromosomes, which appear differentiated from Chipewayan and Mongolian Q-M242* chromosomes. An important differentiation of P-M45* chromosomes between populations is also observed, particularly among Mongolians and Native Americans.
Figure 5.
Neighbor-joining tree relating P-M45 Y lineages on the basis of Nei’s standard genetic distance (Nei 1987), calculated from the allele frequencies observed at the six microsatellite loci examined (table 2). Numbers indicate bootstrap values (%) for 1,000 replicates.
Discussion
The use of genetic polymorphisms to examine the initial peopling of the American continent is complicated by the possibility that some of the diversity observed in present-day populations results from admixture with recent immigrants. This problem could affect Y-chromosome studies more seriously than those based on mtDNA, since there is evidence that admixture in the Americas has preferentially involved immigrant men and native women (Carvajal-Carmona et al. 2000; Mesa et al. 2000). A case in question is the M45 mutation, which is found both in Europeans and Native Americans (fig. 2). Initial Y-chromosome analyses in the Americas placed much emphasis in the possibility of admixture and proposed that P-M45 chromosomes lacking the derived T allele at marker M3 had a recent nonnative origin (Underhill et al. 1996; Bianchi et al. 1997, 1998). The subsequent characterization of Y-microsatellite diversity called into question such an extreme scenario by demonstrating the existence of a subset of P-M45 chromosomes closely related to the Q-M3 haplogroup and pointing to a recent ancestor of Q-M3 as a major founder lineage in the Americas (Ruiz-Linares et al. 1999).
More recently, Lell et al. (2002) proposed the division of P-M45 into two sublineages: M45a, comprising haplotypes closely related to Q-M3; and M45b, comprising haplotypes differentiated from Q-M3 and P-M45 chromosomes carrying the M173 mutation (Lell et al. 2002). Sublineage M45a was observed in southern central Siberia and from North to South America, whereas M45b appears restricted to coastal/eastern Siberia and North America. The contrasting distribution of differentiated Y lineages was interpreted as resulting from two separate male migrations into the Americas (Lell et al. 2002). However, since haplogroup P-M45 is found both in Native Americans and in Europeans, Tarazona-Santos and Santos (2002) challenged this interpretation. These last authors favor a single migration into the continent and propose that the differing distribution of P-M45 haplotypes in the Americas results from variable levels of admixture in North and South American natives.
The M242 mutation recently identified by Seielstad et al. (2003 [in this issue]) is ancestral to M3 within the P-M45 haplogroup and thus allows a refined assessment of P-M45 chromosomes lacking the M3 marker in Native Americans. The Q-M242* linage has a wide distribution in Asia (Seielstad et al. 2003 [in this issue]), and we found it in 8 of the 24 Native American populations examined here, including South Amerinds and the Na Dené sample studied. The M242 marker therefore unambiguously identifies a novel major Native American founder Y lineage ancestral to Q-M3 within the P-M45 haplogroup (fig. 2 and Seielstad et al. 2003 [in this issue]). The characterization of the microsatellite diversity of the Q-M242 haplogroup in Native Americans and Mongolians revealed a close affinity between Amerindian Q-M3 and Q-M242* chromosomes. The modal alleles are the same in both haplogroups (table 2), and network analysis (fig. 4) identified the haplotype carrying the modal alleles at the six loci examined as ancestral for both Q-M3 and Q-M242* chromosomes in Amerindians (haplotypes 5 and 2 in tables 1 and 3, respectively). Thus, the M3 mutation most likely occurred on a Q-M242 chromosome carrying microsatellite alleles 13-12-24-10-14-13 at loci DYS19, DYS388, DYS390, DYS391, DYS392, and DYS393, respectively.
The close relatedness of Amerindian Q-M242* and Q-M3* chromosomes is further illustrated by the neighbor-joining tree relating P-M45 chromosomes (fig. 5). The short distance separating Amerindian Q-M3* and Q-M242* chromosomes reflects the similar microsatellite allele composition of these two haplogroups in Amerinds and substantiates the prior proposal that a fraction of P-M45 chromosomes is closely related to Q-M3 and is native to the Americas (Ruiz-Linares 1999). Q-M242* chromosomes found in Amerinds thus closely match the proposed sublineage M45a of Lell et al. (2002). Microsatellite data also suggest that Q-M242* chromosomes found in the Chipewayan are not closely related to those observed in Amerinds (fig. 5).
In this context, it is interesting to examine the possible origin of P-M45 chromosomes lacking the M242 marker in Native Americans (haplogroup P-M45*). Noticeably, the modal microsatellite haplotype observed in Chipewayan P-M45* chromosomes (haplotype 3 in table 4, with a frequency of ∼29%) includes the most common four-locus haplotype observed in the M45b sublineage of Lell et al. (2002). Tarazona-Santos and Santos (2002) questioned the idea that M45b constitutes a Native American founder lineage, since haplotype 3 (characterized by alleles 14-12-24-11-13-13) is common in European populations (Weale et al. 2002), and its presence in Native Americans could be the result of recent admixture. However, although we did not find this haplotype in Mongolia, closely related lineages (including a one-step neighbor) are present in that region (haplotype 2 in table 4 and fig. 4D). Furthermore, Lell et al. (2002) observed the four-locus equivalent of haplotype 3 (and other closely related haplotypes) in populations from Siberia. The high frequency of haplogroup P-M45* in the Chipewayan (63%; fig. 2) also makes it unlikely that all these chromosomes result from admixture, since such a predominant European ancestry seems inconsistent with the preservation of the cultural identity of this population. A high level of European ancestry in the Chipewayan is also likely to introduce other lineages that are common in Europe (such as DE-YAP* and Y*; fig. 2). However, these were not observed in our sample or in the populations examined by Lell et al. (2002). By comparison, the lineage distribution in South Amerinds is more consistent with a low level of admixture: other than P-M45*, haplogroups Y*, DE-YAP*, and E-M2 (common in Europe and Africa) are also observed in some of the populations examined (fig. 2). Thus, although some of the P-M45* chromosomes found in South Amerinds seem to be of nonnative origin, it is doubtful that the same applies for a large fraction of those observed in the Chipewayan. In fact, the prevalence of major Y lineages in the Chipewayan (including an elevated frequency of P-M45*) is typical of some central Siberian populations (such as the Selkup, Yakut, and Kets), as illustrated by the close affinity of these populations in the principal-component analysis shown in figure 3. Our findings therefore support the proposal of Lell et al. (2002) that a fraction of the haplotype 3–related P-M45* chromosomes observed in Native North Americans originated in Asia and are not the result of recent admixture with Europeans.
Among Native Americans, the RPS4Y711 mutation has so far been reported only in North American populations and in two Wayuu individuals from the extreme north of South America (Karafet et al. 1999; Lell et al. 2002). However, few South American populations have been tested for this polymorphism. In our extensive survey of 23 native South American populations (including an independent sample of Wayuu), the RPS4Y711 mutation was not observed, although we did find it in the North American Chipewayan. These data thus strongly support the notion that the C-RPS4Y711 lineage has a geographic distribution restricted to North America.
Altogether, our findings indicate that Y-chromosome diversity in the Chipewayan contrasts markedly with that observed in South Amerinds, in showing a haplogroup distribution similar to that observed in some central Siberian populations (fig. 3) and the presence of (i) the RPS4Y711 mutation, (ii) differentiated Q-M242* haplotypes, and (iii) P-M45* lineages of Asian origin. The last two observations could underlie the pattern observed by Lell et al. (2002) that led to their proposal of the M45b sublineage. These findings strongly support the notion of a different ancient demographic history for Native North and South Americans and are more simply explained by the recent arrival of new Asian migrants into North America, as suggested by Karafet et al. (1999) and Lell et al. (2002). The lower microsatellite diversity of haplogroups Q-M242* and P-M45* in the Chipewayan relative to South Amerinds could imply a lower Y-chromosome diversity in the Na Dené, consistent with their more recent arrival in the Americas, but this requires confirmation with a more extensive population sampling of the Na Dené.
Noting the high frequency of the C-RPS4Y711 haplogroup in eastern Siberia, Karafet et al. (1999) and Lell et al. (2002) suggested an origin in this region for the second male migration to North America. However, this proposal is not consistent with the generally high frequency of haplogroup K-M9 in eastern Siberia and its absence in the Americas (Karafet et al. 1999; Lell et al. 2002). The principal-component analysis shown in figure 3, in fact, suggests a close genetic relatedness between some Native North Americans (the Chipewayan and the Cheyenne) and certain populations of central/southern Siberia (particularly the Kets, Yakut, Selkup, and Altais), at the resolution of major Y-chromosome haplogroups. This pattern agrees with the distribution of mtDNA haplogroup X, which is found in North America, is absent from eastern Siberia, but is present in the Altais of southern central Siberia (Brown et al. 1998; Smith et al. 1999; Derenko et al. 2001; Malhi and Smith 2002). In agreement with these genetic findings, it has been proposed that Ket and Na Dené languages could be related (Greenberg et al. 1996). These observations suggest that the most recent Asian migration, which appears to have impacted mostly North America, originated in southern/central Siberia. Further evaluation of this hypothesis requires a more detailed characterization of Y-chromosome diversity in central Siberia and North America.
It is interesting to note that haplogroup C-RPS4Y711 and the proposed sublineage M45b have both been detected in North American populations of the Amerind linguistic group (Bergen et al. 1999; Karafet et al. 1999; Lell et al. 2002). The principal-component graph in figure 3 illustrates that the haplogroup distribution in the Chipewayan and the Cheyenne is very similar. At the mtDNA level, it has also been shown that haplogroup X is present both in Na Dené and in North American Amerinds (Brown et al. 1998). These findings do not evidence a close correspondence between the second wave of Asian colonization and the Na Dené linguistic family, as proposed in the model of Greenberg et al. (1986). However, some level of admixture between the Na Dené and North Amerinds is likely and is well documented for the Navajo (Cavalli-Sforza et al. 1994).
Regarding the Asian ancestry of South Amerinds, the high frequency and haplotype diversity of haplogroup P-M45* in southern/central Siberia has led to the proposal of this region as the region of origin for the initial male migration to the Americas (Karafet et al. 1999; Santos et al. 1999; Lell et al. 2002). Consistent with this scenario, the limited microsatellite data available show that Q-M242 chromosomes related to haplotype 13-12-24-10-14-13 (in which the M3 mutation presumably occurred; tables 1 and 3) are found in central Asia, including Mongolia (table 3), and in the Tuvan population (Seielstad et al. 2003 [in this issue]).
Globally, Y-chromosome data therefore emphasize the critical role of southern/central Siberia in the peopling of the Americas, since this region appears to be at the origin of two major male migratory waves of colonization. The data presented here are also consistent with the intriguing possibility of ancient links between proto-Europeans and proto–Native Americans, an idea that has been put forward in previous Y-chromosome studies (Karafet et al. 1999; Santos et al. 1999; Wells et al. 2001; Lell et al. 2002). An ancestral connection between these groups has also been suggested on the basis of morphological (Brace et al. 2001) and mtDNA (Brown et al. 1998) data and could ultimately trace back to ancient east/west human dispersals from a common source in central Asia. In agreement with this scenario, the P-M45 lineage has been found to be oldest in central Asia (Wells et al. 2001; Zerjal et al. 2002), where the Tuvan population includes haplogroups M242, M45, M173, and Tat, which are now dispersed in Europe and/or America.
Lineage Q-M19 had previously been detected in the Ticuna of the upper Amazon and in the Wayuu, a population living in the Guajira peninsula on the extreme north of Colombia’s Caribbean coast (Ruiz-Linares et al. 1999). Both populations have been classified in the Equatorial-Tucano linguistic family (Ruhlen 1991). The Q-M19 lineage was not detected in any of the other South Amerind populations examined here. Our survey included several populations occupying areas intermediate between the Ticuna and the Wayuu and comprises eight additional representatives of the Equatorial-Tucano linguistic family (figs. 1 and 2). The restricted distribution of Q-M19 chromosomes would suggest that this lineage has a recent origin. However, the estimated age for Q-M19 is similar to the one obtained for its parental haplogroup Q-M3 (∼7,000–8,000 ybp). This observation suggests that population isolation and possibly the process of tribalization of Native Americans started soon after the initial human dispersal in the region. Evidence suggestive of an early onset of tribalization in the Americas has also been provided by mtDNA studies that have documented an ancient origin for some private allelic variants (Torroni et al. 1993) and is consistent with the strong genetic drift generally observed in Native American populations (Bortolini et al. 2002). Such an ancient origin for some extant native American populations could be a contributing factor to the great linguistic diversification of Amerinds and their disputed grouping into a single linguistic family (Cavalli-Sforza et al. 1994).
The age calculated for the Q-M242* lineage in Amerinds (13,611 years) and in Mongolia (15,416 years) points to a relatively recent colonization of the Americas. These dates are similar to those obtained with 16 Y-STR markers in a wide sample of Asian Q-M242 chromosomes (Seielstad et al. 2003 [in this issue]). The Asian estimates represent an upper limit for the time of entry into the Americas, and the similarity in age obtained for Q-M242 in Asia and Amerinds is suggestive of an entry soon after the occurrence of this mutation (assuming that entrance in the Americas was associated with an important population bottleneck). Although lineage dating is subject to large error margins, these Y-chromosome estimates are considerably more recent than the ∼20,000–30,000 years calculated for initial entry into America on the basis of mtDNA data (Torroni et al. 1994; Forster et al. 1996; Bonatto and Salzano 1997a, 1997b; Silva et al. 2002). Interestingly, the Y-based dates are comparable to the age estimated for the oldest (well-established) archaeological sites in the Americas (∼12,500 radiocarbon years old [Roosevelt et al. 2002]). Y-chromosome data are thus consistent with a relatively late colonization of the Americas followed by a rapid human population dispersal, as suggested by various archaeological studies (Jablonski 2002).
The discrepancy between the Y-chromosome and mtDNA estimates for the time of initial colonization could have several explanations, of which only two are briefly mentioned here. It could relate to a more severe contraction of the male population at the time of initial entry into the Americas. Critically, migration age estimates assume the introduction of a single haplotype per lineage at the time of initial colonization. However, there is growing evidence that founder heterogeneity exists within the four major recognized Native American mtDNA lineages (Malhi et al. 2002). Another factor that could contribute to a Y-chromosome/mtDNA discrepancy is the inaccuracy of mutation rate estimates—particularly, the direct observation of mutations in mother-offspring pairs suggests that the mutation rate of mtDNA could be considerably higher than the molecular clock calibrations usually employed in population studies (Heyer et al. 2001). Both a failure to account for mtDNA founder heterogeneity and an underestimation of the mutation rate for mtDNA would increase the calculated date of initial entry into the Americas (Weiss 1994).
Acknowledgments
We are very grateful to the individuals who donated the samples analyzed here. Some of these samples were collected in collaboration with Professors F. M. Salzano, Francis L. Black, and the late James V. Neel. Thanks are due to the Brazilian Fundação Nacional do Índio for logistic support. This investigation was approved by the Brazilian National Ethics Commission, the Canadian Institutional Review Boards of the Sainte-Justine (Montreal) and Victoria (Prince Albert) Hospitals, The Prince Albert Grand Council, and the Bioethics Committee of Universidad de Antioquia (Colombia). Financial support in Brazil was provided by PRONEX, CNPq, and the Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul. Laboratory work in the United Kingdom was funded by a Wellcome Trust Travelling Research Fellowship (grant 059763 to M.-C.B. and A.R.-L.). D.L. is supported by Canadian Institutes of Health Research grant MOP-12782. We are grateful to Mark Seielstad and Peter Underhill, for information on the M242 polymorphism; to T. A. Weimer, for encouragement; and to two anonymous reviewers, for their insightful comments on this work. We also thank Oliver Burbage-Hall, Zara Qadir, and Ratna Kundi, for technical assistance, and Alisa Izraeljan, for help in the preparation of the manuscript.
References
- Bandelt HJ, Forster P, Sykes BC, Richards MB (1995) Mitochondrial portraits of human populations using median networks. Genetics 141:743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen AW, Wang CY, Tsai J, Jefferson K, Dey C, Smith KD, Park SC, Tsai SJ, Goldman D (1999) An Asian-Native American paternal lineage identified by RPS4Y resequencing and by microsatellite haplotyping. Ann Hum Genet 63:63–80 [DOI] [PubMed] [Google Scholar]
- Bianchi N, Bailliet G, Brave CM, Pena SD, Rothhammer F (1997) Origin of Amerindian Y-chromosome as inferred by the analysis of six polymorphic markers. Am J Phys Anthropol 102:79–89 [DOI] [PubMed] [Google Scholar]
- Bianchi NO, Catanesi CI, Bailliet G, Martinez-Marignac VL, Bravi CM, Vidal-Rioja LB, Herrera RJ, Lopez-Camelo JS (1998) Characterization of ancestral and derived Y-chromosome haplotypes of New World native populations. Am J Hum Genet 63:1862–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonatto SL, Salzano FM (1997a) Diversity and age of the four major mtDNA haplogroups, and their implications for the peopling of the New World. Am J Hum Genet 61:1413–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (1997a) A single and early migration for the peopling of the Americas supported by mitochondrial DNA sequence data. Proc Natl Acad Sci USA 94:1866–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolini MC, Salzano FM, Bau CH, Layrisse Z, Petzl-Erler ML, Tsuneto LT, Hill K, Hurtado AM, Castro-De-Guerra D, Bedoya G, Ruiz-Linares A (2002) Y-chromosome biallelic polymorphisms and Native American population structure. Ann Hum Genet 66:255–259 [DOI] [PubMed] [Google Scholar]
- Brace CL, Nelson AR, Seguchi N, Oe H, Sering L, Qifeng P, Yongyi L, Tumen D (2001) Old World sources of the first New World human inhabitants: a comparative craniofacial view. Proc Natl Acad Sci USA 98:10017–10022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MD, Hosseini SH, Torroni A, Bandelt HJ, Allen JC, Schurr TG, Scozzari R, Cruciani F, Wallace DC (1998) mtDNA haplogroup X: an ancient link between Europe/Western Asia and North America? Am J Hum Genet 63:1852–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal-Carmona LG, Soto ID, Pineda N, Ortiz-Barrientos D, Duque C, Ospina-Duque J, McCarthy M, Montoya P, Alvarez VM, Bedoya G, Ruiz-Linares A (2000) Strong Amerind/white sex bias and a possible Sephardic contribution among the founders of a population in northwest Colombia. Am J Hum Genet 67:1287–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A (1994) The history and geography of human genes. Princeton University Press, Princeton [Google Scholar]
- Crawford MH (1998) The origins of Native Americans. Cambridge University Press, Cambridge [Google Scholar]
- Derenko MV, Grzybowski T, Malyarchuk BA, Czarny J, Miscicka-Sliwka D, Zaharov IA (2001) The presence of mitochondrial haplogroup X in Altaians from South Siberia. Am J Hum Genet 69:237–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J (2001) PHYLIP: phylogeny inference package. University of Washington, Seattle [Google Scholar]
- Fiedel SJ (1992) Prehistory of the Americas. Cambridge University Press, Cambridge [Google Scholar]
- Forster P, Harding R, Torroni A, Bandelt HJ (1996) Origin and evolution of Native American mtDNA variation: a reappraisal. Am J Hum Genet 59:935–945 [PMC free article] [PubMed] [Google Scholar]
- Goldstein DB, Ruiz Linares A, Cavalli-Sforza LL, Feldman MW (1995) An evaluation of genetic distances for use with microsatellite loci. Genetics 139:463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg JH (1996) The “Greenberg” hypothesis. Science 274:1447 [DOI] [PubMed] [Google Scholar]
- Greenberg JH, Turner CG, Zegura SL (1986) The settlement of the Americas: a comparison of the linguistic, dental and genetic evidence. Curr Anthropol 27:477–497 [Google Scholar]
- Heyer E, Zietkiewicz E, Rochowski A, Yotova V, Puymirat J, Labuda D (2001) Phylogenetic and familial estimates of mitochondrial substitution rates: study of control region mutations in deep-rooting pedigrees. Am J Hum Genet 69:1113–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horai S, Kondo R, Nakagawa-Hattori Y, Hayashi S, Sonoda S, Tajima K (1993) Peopling of the Americas, founded by four major lineages of mitochondrial DNA. Mol Biol Evol 10:23–47 [DOI] [PubMed] [Google Scholar]
- Jablonski NG (ed) (2002) The first Americans: the Pleistocene colonization of the New World. California University Press, San Francisco [Google Scholar]
- Karafet TM, Zegura SL, Posukh O, Osipova L, Bergen A, Long J, Goldman D, Klitz W, Harihara S, de Knijff P, Wiebe V, Griffiths RC, Templeton AR, Hammer MF (1999) Ancestral Asian source(s) of new world Y-chromosome founder haplotypes. Am J Hum Genet 64:817–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karafet T, Zegura SL, Vuturo-Brady J, Posukh O, Osipova L, Wiebe V, Romero F, Long JC, Harihara S, Jin F, Dashnyam B, Gerelsaikhan T, Omoto K, Hammer MF (1997) Y chromosome markers and trans-Bering Strait dispersals. Am J Phys Anthropol 102:301–314 [DOI] [PubMed] [Google Scholar]
- Kayser M, Roewer L, Hedman M, Henke L, Henke J, Brauer S, Kruger C, Krawczak M, Nagy M, Dobosz T, Szibor R, de Knijff P, Stoneking M, Sajantila A (2000) Characteristics and frequency of germline mutations at microsatellite loci from the human Y chromosome, as revealed by direct observation in father/son pairs. Am J Hum Genet 66:1580–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolman CJ, Sambuughin N, Bermigham E (1996) Mitochondrial DNA analysis of Mongolian populations and implications for the origin of new world founders. Genetics 142:1321–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lell JT, Brown MD, Schurr TG, Sukernik RI, Starikovskaya YB, Torroni A, Moore LG, Troup GM, Wallace DC (1997) Y chromosome polymorphisms in Native American and Siberian populations: identification of Native American Y chromosome haplotypes. Hum Genet 100:536–543 [DOI] [PubMed] [Google Scholar]
- Lell JT, Sukernik RI, Starikovskaya YB, Su B, Jin L, Schurr TG, Underhill PA, Wallace DC (2002) The dual origin and Siberian affinities of Native American Y chromosomes. Am J Hum Genet 70:192–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhi RS, Eshleman JA, Greenberg JA, Weiss DA, Schultz-Shook BA, Kaestle FA, Lorenz JG, Kemp BM, Johnson JR, Smith DG (2002) The structure of diversity within New World mitochondrial DNA haplogroups: implications for the prehistory of North America. Am J Hum Genet 70:905–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhi RS, Smith DG (2002) Brief communication: haplogroup X confirmed in prehistoric North America. Am J Phys Anthropol 119:84–86 [DOI] [PubMed] [Google Scholar]
- Merriwether DA, Ferrell RE (1996) The four founding lineage hypothesis for the New World: a critical reevaluation. Mol Phylogenet Evol 5:241–246 [DOI] [PubMed] [Google Scholar]
- Merriwether DA, Hall WW, Vahlne A, Ferrell RE (1996) mtDNA variation indicates Mongolia may have been the source for the founding population for the New World. Am J Hum Genet 59:204–212 [PMC free article] [PubMed] [Google Scholar]
- Mesa NR, Mondragon MC, Soto ID, Parra MV, Duque C, Ortiz-Barrientos D, Garcia LF, Velez ID, Bravo ML, Munera JG, Bedoya G, Bortolini MC, Ruiz-Linares A (2000) Autosomal, mtDNA, and Y-chromosome diversity in Amerinds: pre- and post-Colombian patterns of gene flow in South America. Am J Hum Genet 67:1277–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel JV, Biggar RJ, Sukernik RI (1994) Virologic and genetic studies relate Amerind origins to the indigenous people of the Mongolia/Manchuria/southeastern Siberia region. Proc Natl Acad Sci USA 91:10737–10741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York [Google Scholar]
- Pena SD, Santos FR, Bianchi NO, Bravi CM, Carnese FR, Rothhammer F (1995) A major founder Y-chromosome haplotype in Amerindians. Nat Genet 11:15–16 [DOI] [PubMed] [Google Scholar]
- Rohlf J (2001) Ntsys: numerical taxonomy and multivariate analysis system version 2.1. Exeter Software, Setauket [Google Scholar]
- Roosevelt AC, Douglas J, Brown L (2002) The migrations and adaptations of the first Americans: Clovis and pre-Clovis viewed from South America. In: Jablonski NG (ed) The first Americans: the Pleistocene colonization of the New World. California University Press, San Francisco, pp 159–235 [Google Scholar]
- Ruhlen M (1991) A guide to the world’s languages. Stanford University Press, Stanford [Google Scholar]
- Ruiz-Linares A, Ortiz-Barrientos D, Figueroa M, Mesa N, Munera JG, Bedoya G, Velez ID, Garcia LF, Perez-Lezaun A, Bertranpetit J, Feldman MW, Goldstein DB (1999) Microsatellites provide evidence for Y chromosome diversity among the founders of the New World. Proc Natl Acad Sci USA 96:6312–6317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos FR, Bianchi NO, Pena SD (1996) Worldwide distribution of human Y-chromosome haplotypes. Genome Res 6:601–611 [DOI] [PubMed] [Google Scholar]
- Santos FR, Pandya A, Tyler-Smith C, Pena SD, Schanfield M, Leonard WR, Osipova L, Crawford MH, Mitchell RJ (1999) The central Siberian origin for native American Y chromosomes. Am J Hum Genet 64:619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S, Roessli D, Excoffier L (2000) Arlequin version 2000: a software for population genetic data analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland [Google Scholar]
- Seielstad M, Yuldasheva N, Singh N, Underhill P, Oefner P, Shen P, Wells RS (2003) A novel Y-chromosome variant puts an upper limit on the timing of first entry into the Americas. Am J Hum Genet 73:700–705 (in this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva WA Jr, Bonatto SL, Holanda AJ, Ribeiro-Dos-Santos AK, Paixao BM, Goldman GH, Abe-Sandes K, Rodriguez-Delfin L, Barbosa M, Paco-Larson ML, Petzl-Erler ML, Valente V, Santos SE, Zago MA (2002) Mitochondrial genome diversity of Native Americans supports a single early entry of founder populations into America. Am J Hum Genet 71:187–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DG, Malhi RS, Eshleman J, Lorenz JG, Kaestle FA (1999) Distribution of mtDNA haplogroup X among Native North Americans. Am J Phys Anthropol 110:271–284 [DOI] [PubMed] [Google Scholar]
- Tarazona-Santos E, Santos FR (2002) The peopling of the Americas: a second major migration? Am J Hum Genet 70:1377–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MG, Bradman N, Flinn HM (1999) High throughput analysis of 10 microsatellite and 11 diallelic polymorphisms on the human Y-chromosome. Hum Genet 105:577–581 [DOI] [PubMed] [Google Scholar]
- Torroni A, Neel JV, Barrantes R, Schurr TG, Wallace DC (1994) Mitochondrial DNA “clock” for the Amerinds and its implications for timing their entry into North America. Proc Natl Acad Sci USA 91:1158–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Schurr TG, Cabell MF, Brown MD, Neel JV, Larsen M, Smith DG, Vullo CM, Wallace DC (1993) Asian affinities and continental radiation of the four founding Native American mtDNAs. Am J Hum Genet 53:563–590 [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Schurr TG, Yang CC, Szathmary EJ, Williams RC, Schanfield MS, Troup GA, Knowler WC, Lawrence DN, Weiss KM (1992) Native American mitochondrial DNA analysis indicates that the Amerind and the Nadene populations were founded by two independent migrations. Genetics 130:153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Jin L, Zemans R, Oefner PJ, Cavalli-Sforza LL (1996) A pre-Colombian Y chromosome-specific transition and its implications for human evolutionary history. Proc Natl Acad Sci USA 93:196–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Passarino G, Lin AA, Shen P, Mirazon LM, Foley RA, Oefner PJ, Cavalli-Sforza LL (2001) The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet 65:43–62 [DOI] [PubMed] [Google Scholar]
- Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonne-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi SQ, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza LL, Oefner PJ (2000) Y chromosome sequence variation and the history of human populations. Nat Genet 26:358–361 [DOI] [PubMed] [Google Scholar]
- Weale ME, Weiss DA, Jager RF, Bradman N, Thomas MG (2002) Y chromosome evidence for Anglo-Saxon mass migration. Mol Biol Evol 19:1008–1021 [DOI] [PubMed] [Google Scholar]
- Weiss KM (1994) American origins. Proc Natl Acad Sci USA 91:833–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells RS, Yuldasheva N, Ruzibakiev R, Underhill PA, Evseeva I, Blue-Smith J, Jin L, et al (2001) The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci USA 98:10244–10249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Y Chromosome Consortium (2002) A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 12:339–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerjal T, Wells RS, Yuldasheva N, Ruzibakiev R, Tyler-Smith C (2002) A genetic landscape reshaped by recent events: Y-chromosomal insights into central Asia. Am J Hum Genet 71:466–482 [DOI] [PMC free article] [PubMed] [Google Scholar]