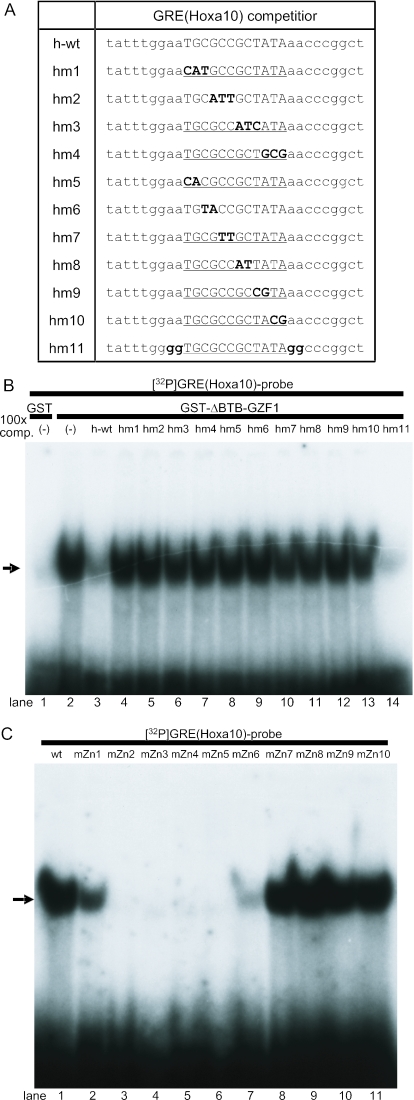
Figure 8.
Mutation analysis of the GRE(HOXA10) sequence and zinc finger motifs in GZF1 required for DNA–protein complex formation. (A) Oligonucleotide sequences used for the competitive EMSAs. Hm1–hm4 contain triple base substitutions in the wild-type (h-wt) GRE(HOXA10) sequence, andhm5–hm10 contain double base substitutions. Hm11 mutated outside ofGRE(HOXA10) is also used in this assay. (B) 100-fold molar excess of unlabeled probes was added to the binding reaction as cold competitors. The wild-type and hm11 oligonucleotides efficiently competed for GST-ΔBTB-GZF1 binding to 32P-labeled GRE(HOXA10), whereas hm1–hm10 cold competitors did not compete at all. An arrow indicates the DNA–protein complex. (C) EMSA using 32P-labeled GRE(HOXA10) and GST-ΔBTB-GZF1 mutated in the individual zinc finger motifs. The arrow indicates the DNA–protein complex.