Abstract
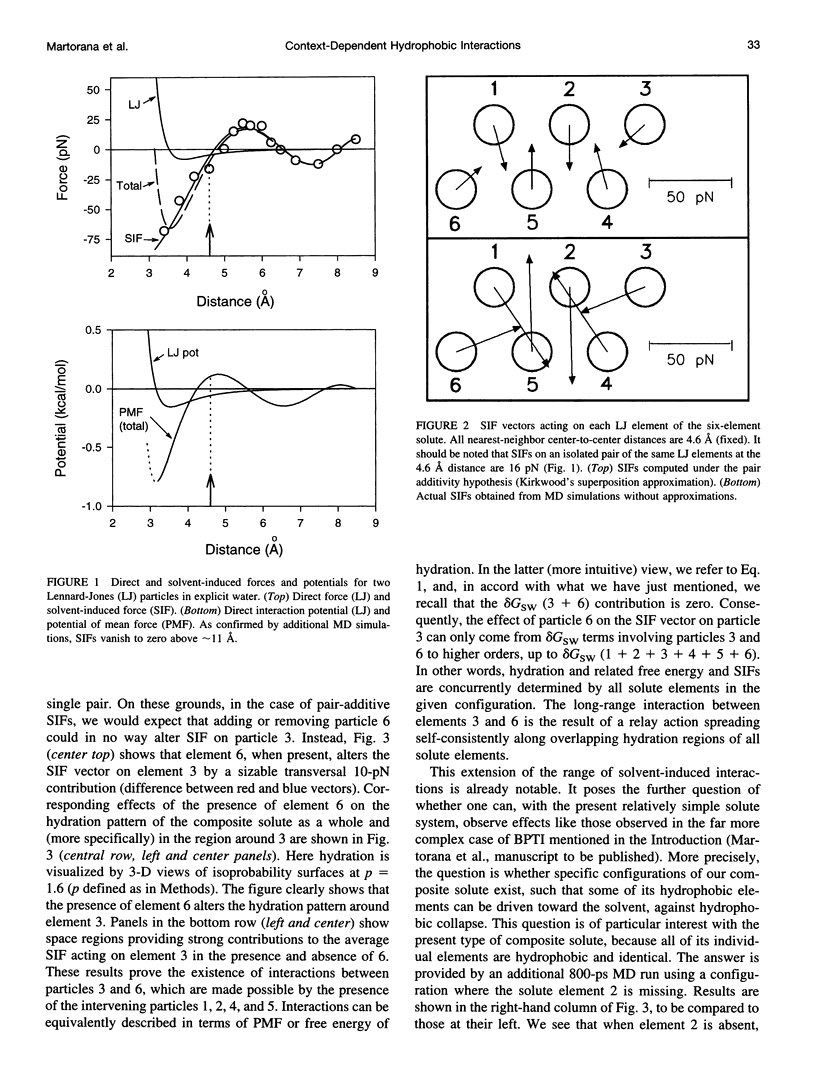
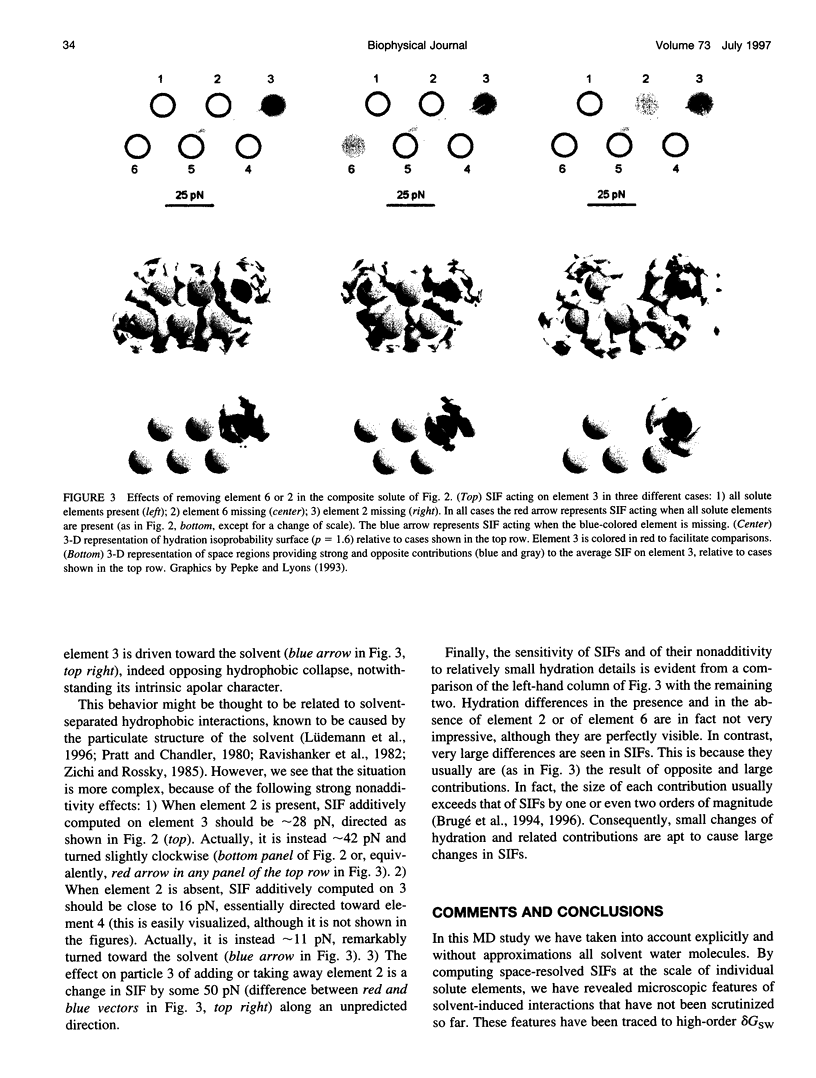
We report results of molecular dynamics (MD) simulations of composite model solutes in explicit molecular water solvent, eliciting novel aspects of the recently demonstrated, strong many-body character of hydration. Our solutes consist of identical apolar (hydrophobic) elements in fixed configurations. Results show that the many-body character of PMF is sufficiently strong to cause 1) a remarkable extension of the range of hydrophobic interactions between pairs of solute elements, up to distances large enough to rule out pairwise interactions of any type, and 2) a SIF that drives one of the hydrophobic solute elements toward the solvent rather than away from it. These findings complement recent data concerning SIFs on a protein at single-residue resolution and on model systems. They illustrate new important consequences of the collective character of hydration and of PMF and reveal new aspects of hydrophobic interactions and, in general, of SIFs. Their relevance to protein recognition, conformation, function, and folding and to the observed slight yet significant nonadditivity of functional effects of distant point mutations in proteins is discussed. These results point out the functional role of the configurational and dynamical states (and related statistical weights) corresponding to the complex configurational energy landscape of the two interacting systems: biomolecule + water.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ben-Naim A. Solvent effects on protein association and protein folding. Biopolymers. 1990 Feb 15;29(3):567–596. doi: 10.1002/bip.360290312. [DOI] [PubMed] [Google Scholar]
- Bulone D., San Biagio P. L., Palma-Vittorelli M. B., Palma M. U. The role of water in hemoglobin function and stability. Science. 1993 Feb 26;259(5099):1335–1336. doi: 10.1126/science.8446903. [DOI] [PubMed] [Google Scholar]
- Chothia C., Wodak S., Janin J. Role of subunit interfaces in the allosteric mechanism of hemoglobin. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3793–3797. doi: 10.1073/pnas.73.11.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill K. A., Bromberg S., Yue K., Fiebig K. M., Yee D. P., Thomas P. D., Chan H. S. Principles of protein folding--a perspective from simple exact models. Protein Sci. 1995 Apr;4(4):561–602. doi: 10.1002/pro.5560040401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garde S, Hummer G, García AE, Pratt LR, Paulaitis ME. Hydrophobic hydration: Inhomogeneous water structure near nonpolar molecular solutes. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 1996 May;53(5):R4310–R4313. doi: 10.1103/physreve.53.r4310. [DOI] [PubMed] [Google Scholar]
- Green S. M., Shortle D. Patterns of nonadditivity between pairs of stability mutations in staphylococcal nuclease. Biochemistry. 1993 Sep 28;32(38):10131–10139. doi: 10.1021/bi00089a032. [DOI] [PubMed] [Google Scholar]
- KAUZMANN W. Some factors in the interpretation of protein denaturation. Adv Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- Klement R., Soumpasis D. M., Jovin T. M. Computation of ionic distributions around charged biomolecular structures: results for right-handed and left-handed DNA. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4631–4635. doi: 10.1073/pnas.88.11.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis T., Archontis G., Karplus M. Enthalpic contribution to protein stability: insights from atom-based calculations and statistical mechanics. Adv Protein Chem. 1995;47:231–306. doi: 10.1016/s0065-3233(08)60547-1. [DOI] [PubMed] [Google Scholar]
- LiCata V. J., Ackers G. K. Long-range, small magnitude nonadditivity of mutational effects in proteins. Biochemistry. 1995 Mar 14;34(10):3133–3139. doi: 10.1021/bi00010a001. [DOI] [PubMed] [Google Scholar]
- San Biagio P. L., Bulone D., Emanuele A., Palma M. U. Self-assembly of biopolymeric structures below the threshold of random cross-link percolation. Biophys J. 1996 Jan;70(1):494–499. doi: 10.1016/S0006-3495(96)79595-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Biagio P. L., Palma M. U. Spinodal lines and Flory-Huggins free-energies for solutions of human hemoglobins HbS and HbA. Biophys J. 1991 Aug;60(2):508–512. doi: 10.1016/S0006-3495(91)82078-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciortino F., Prasad K. U., Urry D. W., Palma M. U. Self-assembly of bioelastomeric structures from solutions: mean-field critical behavior and Flory-Huggins free energy of interactions. Biopolymers. 1993 May;33(5):743–752. doi: 10.1002/bip.360330504. [DOI] [PubMed] [Google Scholar]



