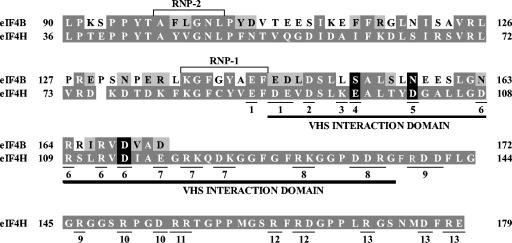
FIG. 6.
Residues of eIF4H whose alteration reduces Vhs binding. The region of sequence homology shared by eIF4H and eIF4B is shown, with the eIF4H amino acids and those eIF4B residues that are identical to amino acids in eIF4H indicated by white letters on a dark grey background. eIF4B residues that represent conservative changes relative to those in eIF4H are depicted by black letters on a light grey background. eIF4B residues that are nonconservative changes from eIF4H amino acids are indicated by black letters on a white background. eIF4H and eIF4B share an RNA recognition motif RNA-binding domain containing two RNP motifs (RNP-1 and RNP-2) that are boxed. The Vhs interaction domain of eIF4H, as defined by deletion mutagenesis (18), is indicated by the dark line under the eIF4H sequence. Thirteen eIF4H mutants were constructed in which clusters of charged amino acids were altered by site-directed mutagenesis. The amino acids that were altered in a particular mutant are underlined and indicated by the number of the mutant under the residue. For example, in mutant 1, E88, D90, and E91 were altered together. All of the indicated charged amino acids were changed to alanine except for mutant 8, in which R139 and K130 were both changed to threonine. The three charged eIF4H amino acids, whose modification to alanine significantly reduces Vhs binding, are indicated by white type on a black background, as are the corresponding residues of eIF4B.