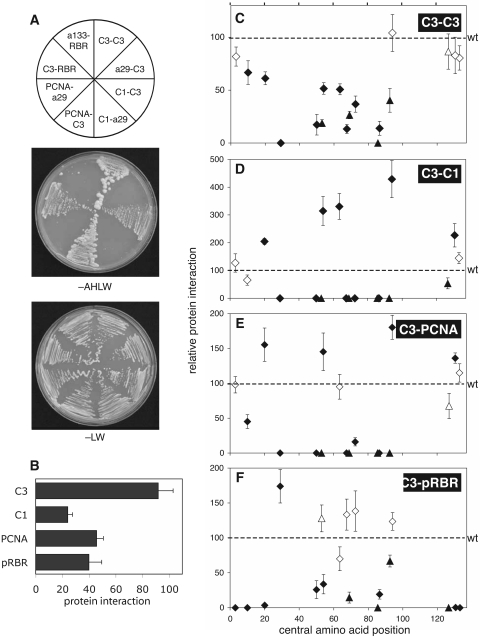
FIG. 4.
TYLCV C3 protein interactions. (A) C3 protein interactions were measured in yeast two-hybrid assays. Wild-type or mutated C3 sequence was fused to the GAL4 activation domain (indicated by the first protein listed in each protein pair) or the GAL4 DNA binding domain (second protein listed). Transformants were selected for protein interactions on medium lacking adenine, histidine, leucine, and tryptophan (−AHLW). Medium lacking leucine and tryptophan (−LW) selected for the input plasmids only. Yeast cotransfected with a wild-type C3 cassette and a cassette corresponding to C3, C1, PCNA, or pRBR grew on both −AHLW and −LW media. Yeast cotransfected with certain mutant C3 cassettes and a C3, C1, PCNA, or pRBR cassette grew only on −LW medium. (B) The strength of the interactions shown for wild-type C3 in panel A was quantified in growth assays. Yeast growth on −AHLW medium was normalized to growth on −LW medium as a measure of strength. (C to F) Growth assays for yeast cotransfected with indicated C3 mutants and (C) wild-type C3, (D) C1, (E) PCNA, and (F) pRBR. Growth in the presence of mutated C3 proteins was normalized to growth in the presence of wild-type C3, which was set at 100. Proteins that were not significantly different than wild type, determined as in Fig. 3, are shown as open symbols. Significantly different proteins are marked with filled symbols. Each data point reflects at least four independent experiments, with the bars representing 2 standard errors. The diamonds and triangles are defined in the Fig. 2 legend.