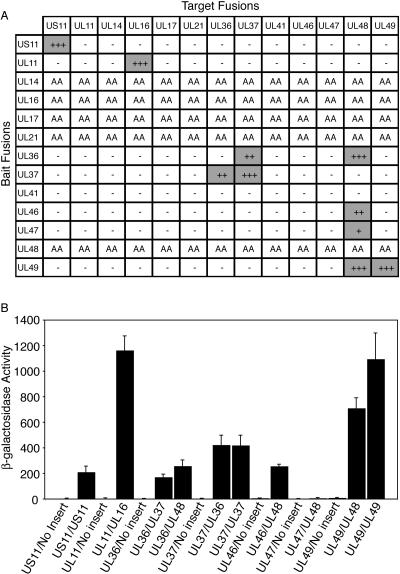
FIG. 1.
Yeast two-hybrid analysis of the interaction of HSV-1 tegument proteins. (A) Summary of data obtained with yeast cotransformed with various combinations of bait and target fusion constructs. Interactions were initially assessed using a qualitative in vivo plate assay (at day 3), with readout being expression of the reporter genes LEU2 and lacZ. Positive interactions (shaded) were observed as blue colonies, with + indicating the lightest blue and +++ indicating the darkest blue. No growth (−) indicates a negative interaction. Autoactivation (AA) was observed for a number of bait constructs. (B) Summary of quantitative liquid β-galactosidase assay. The activity was calculated from the following equation: β-galactosidase activity = 1,000 × A420/(t × V × OD660), where t is time (in minutes) of incubation, V is volume of cells (in ml) used in the assay, and OD660 is the optical density at 660 nm. The values obtained for β-galactosidase activity are the averages of measurements from at least three separate colonies. Negative controls consisted of displayBait/viral gene insert and displayTarget/no insert. UL36 in displayBait encodes amino acids 1 to 1874, while UL36 in displayTarget encodes amino acids 1 to 767.