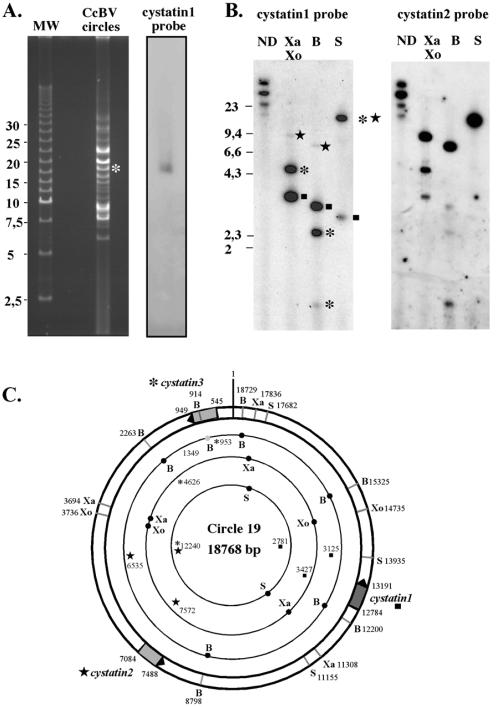
FIG. 2.
Mapping of Cotesia congregata bracovirus cystatins in the viral genome. (A) Field inversion gel electrophoresis of 250 ng of viral DNA, followed by hybridization using cystatin 1 cDNA as a probe. The unique hybridization signal observed corresponds to the circle (indicated by a white asterisk) of the estimated linear size (19 kb). The 2.5-kb ladder from Bio-Rad was used as linear molecular weight marker in this experiment. (B) Southern blot analysis of viral DNA (200 ng per lane), nondigested (ND) or after restriction digestion with XbaI-XhoI (Xa, Xo), BamHI (B), or SacI (S), using either cystatin 1 cDNA or cystatin 2 cDNA as a probe. Hybridizing fragments corresponding to cystatin 1 are indicated by a black square. Hybridizing fragments corresponding to cystatin 2 are indicated by a black star. Hybridizing fragments corresponding to cystatin 3 are indicated by a black asterisk. (C) Restriction map of CcBV circle19, harboring the three cystatin genes. Lengths of expected fragments after restriction digestion are indicated. Hybridizing fragments are highlighted with corresponding symbols (box, star, or asterisk). Note the presence of a BamHI site (grey circle) in cystatin 3, which gives rise to a partial digest.