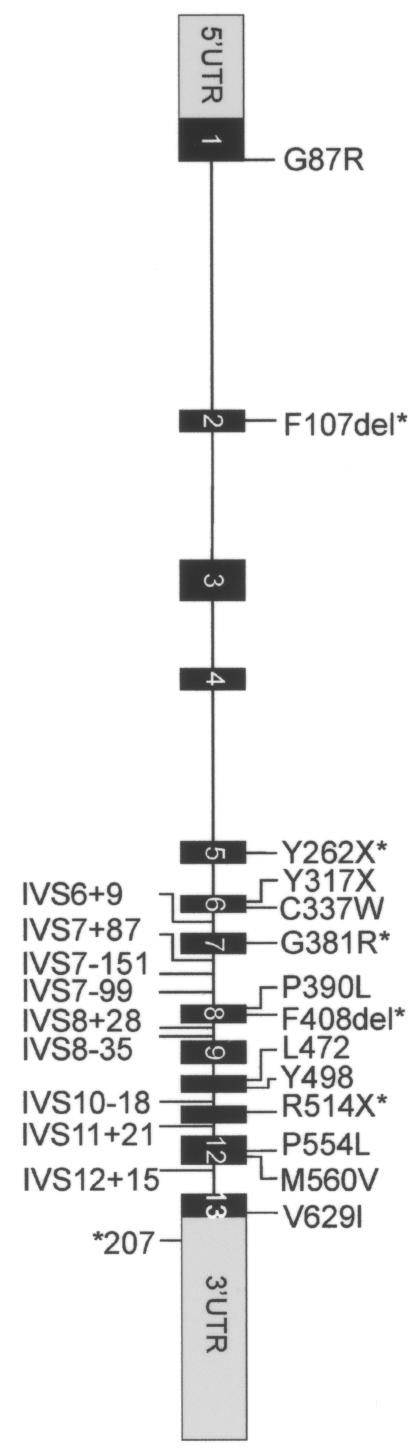
Figure 1.
Mutations and polymorphisms in patients with XLMR. Primer sequences were designed specifically to amplify all exons of the SLC6A8 gene (and not the SLC6A10 gene, a presumed creatine transporter pseudogene mapped at chromosome 16 [Eichler et al. 1996]), including short fragments of the flanking intronic sequences. By comparing the SLC6A8 and SLC6A10 sequences, we have found that at least two nucleotide variations were present in all amplicons, confirming selective amplification of the SLC6A8 sequences (data not shown). PCR reactions were performed with HotStar Taq (Qiagen) in a PE Applied Biosystems model 9700. Direct sequence analysis was performed on purified PCR products (Millipore vacufold) by use of BigDye v3.1 terminators and an ABI 3100 sequence machine (PE Applied Biosystems). The obtained electropherograms were assembled and analyzed to identify potential genomic alterations by use of the Mutation Surveyor software package (SoftGenetics). Sequence variants were annotated according the guidelines of den Dunnen and Antonarakis (2001). On the basis of impaired uptake in fibroblasts, five alterations (asterisks [*]) have been proven elsewhere to be mutations. p.F107del was also found in our XLMR cohort. One novel nonsense mutation (p.Y317X) is strongly predictive of impaired creatine uptake. p.M560V is a rare polymorphism, and p.V629I is an unclassified variant. The implications of the translational silent variant, the IVS variant, and the 3′ UTR variant could not yet be investigated, and these variants are therefore assigned as “unclassified.” IVS variants in introns 1, 2, 3, and 4 may have been missed, since only small exon-flanking parts were included.