Abstract
The IL12B gene on chromosome 5q31-33 encodes the p40 subunit of interleukin 12, an immunomodulatory cytokine. To test the hypothesis that the IL12B gene contains polymorphisms associated with asthma, we genotyped six haplotype-tagging polymorphisms in the IL12B gene, both in 708 children enrolled in the Childhood Asthma Management Program (CAMP) and in their parents. Using the family-based association test (FBAT) program and its haplotype (HBAT) and phenotype (PBAT) options, we tested each polymorphism and haplotype for association with asthma and asthma-related phenotypes. We tested positive associations for replication in a case-control study comparing 177 adult moderate-to-severe asthmatics with 177 nonasthmatic controls. In whites in the CAMP cohort, the A allele of the IL12B G4237A polymorphism was undertransmitted to asthmatic children (P=.0008, recessive model), the global test for haplotypes for affection status was positive (P=.009, multiallelic χ2), and two polymorphisms were associated with different atopy phenotypes. In addition, we found a strong association between the IL12B_4237 and IL12B_6402 polymorphisms and an asthma-severity phenotype in whites, which we also found in the independent population of white adult asthmatics. IL12B may be an important asthma gene.
Chromosome 5q31 contains numerous immunoregulatory genes that could influence asthma and atopy phenotypes and that has been linked to those phenotypes in many genetic studies (Meyers et al. 1994; Rosenwasser et al. 1995; Cookson and Moffatt 2000; Walley et al. 2001). Interleukin 12B (IL12B [MIM 161561]) is located in this genomic region and encodes the 40-kDa subunit of the heterodimeric glycoprotein that is IL12 (Huang et al. 2000). IL12 is an immunomodulatory cytokine that is the primary inducer of the development of T-helper 1 (Th1) cells, with downregulation of T-helper 2 (Th2) cytokines that are associated with asthma (Trinchieri and Scott 1994). IL12 also plays a major role in the mammalian innate immune response to viruses, a major trigger for asthma induction (Yap et al. 2000). Therefore, IL12B is a strong biological candidate gene for asthma. Shikano et al. (2001) reported possible abnormalities in the IL12 signaling pathways in atopic patients; however, Noguchi et al. (2001) did not find any association between polymorphisms in IL12B and atopy phenotypes. Morahan and colleagues (2002) reported an association between heterozygosity for an IL12B-promoter polymorphism and asthma severity in atopic and nonatopic individuals. Our objective was to investigate associations between polymorphisms in IL12B and mild-to-moderate asthma by use of a family-based approach, with confirmation by use of a population approach in the same family sample and in an independent adult sample.
The characteristics of both the pediatric and adult populations are shown in table 1. The Childhood Asthma Management Program (CAMP) was a multicenter, randomized, double-masked, placebo-controlled clinical trial that evaluated the long-term effects of inhaled anti-inflammatory medications in children with mild-to-moderate asthma (Childhood Asthma Management Program Research Group 1999). The diagnosis of asthma was based on one or more of the following criteria for at least 6 mo in the year prior to recruitment: (1) asthma symptoms at least two times per wk, (2) at least two usages per wk of an inhaled bronchodilator, and (3) daily asthma medication. Methacholine hyperreactivity, as indicated by the dose of the drug that caused a 20% decrease in forced expiratory volume at 1 s (FEV1) was ⩽12.5 mg/ml. Spirometry was performed according to the American Thoracic Society recommendations, by use of a volume-displacement spirometer, and airway responsiveness was assessed by methacholine challenge, by use of the Wright nebulizer tidal-breathing technique, during a baseline pretrial period in which children had been medication-free for at least 28 d (Childhood Asthma Management Program Research Group 1999). Serum eosinophil counts were performed at each enrolling center. Total serum immunoglobulin E (IgE) was measured using radioimmunoabsorbent assays from blood samples collected during the screening sessions.
Table 1.
Characteristics of Study Population
|
Adults |
|||
| Characteristic | CAMP Asthmatic Children | Asthmatics | Controls |
| Population size | N=708 | N=177 | N=177 |
| % male | 59.4% | 45 | 62 |
| Age in years (SD) | 8.07 (±2.1) | 33.7 (±13.7) | 25.7 (±7.6) |
| Ethnicity (%): | |||
| Non-Hispanic white | 73.4 | 100 | 100 |
| African American | 9.7 | 0 | 0 |
| Hispanic | 7.5 | 0 | 0 |
| Other | 9.2 | 0 | 0 |
| Baseline FEV1% predicted (SD) | 95.1 (±13) | 61.8 (±10.5) | 101.2 (±14.6) |
DNA samples were obtained from 968 children (of 1,041 children who participated in the original CAMP study) (The Childhood Asthma Management Program Research Group 2000) and from 1,518 of their parents. Complete family trios were available for 652 nuclear families, which included 708 children. Some families had multiple children with asthma (49 families had two children, and 2 families had three children). The characteristics of these families are shown in table 1.
The adult asthmatics and controls were all white. The adult cases included 177 patients with asthma who were originally recruited for an asthma-medication trial in the United States. To qualify for inclusion, patients had to be nonsmokers aged 18–45 years, have no significant comorbid medical conditions, and have diagnostic findings consistent with moderate-to-severe asthma, according to the American Thoracic Society criteria (American Thoracic Society 1987). Patients had an FEV1 of 40%–85% of the predicted normal values (Crapo et al. 1982) after at least 8 h of not inhaling beta agonist medications and after receiving no corticosteroids for at least 6 wk. A ⩾15% change in FEV1 after beta-agonist or methacholine-sensitivity testing was used to confirm asthma diagnosis. The adult control population included DNA from 177 subjects without asthma from the Environmental Medicine Genome Bank (Sonna et al. 2000). This DNA bank consists of DNA collected from U.S. Army recruits from across the country during basic training: Army recruits are screened to exclude those who have a history of asthma or obstructive airway disease. The characteristics of the cases and controls are shown in table 1.
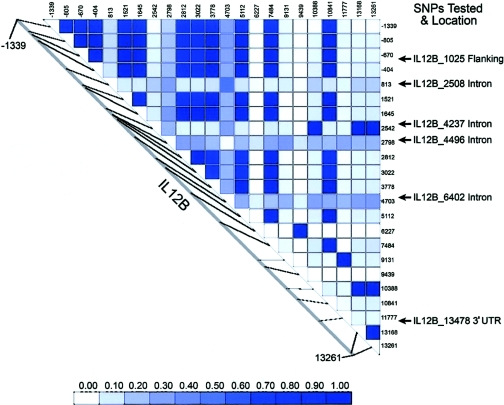
Because 73.4% of the sample was white, SNPs were chosen to ensure full representation of the gene, on the basis of haplotypes inferred using the PHASE software (Stephens et al. 2001), from resequencing data reported on the University of Washington–Fred Hutchinson Cancer Research Center (UW-FHCRC) Web site (accessed December 2002). These data were derived from 23 white individuals who were CEPH parents (DNA from the Coriell Cell Repository). Haplotypes were inferred using 21 SNPs, for which the minor allele was at a frequency of ⩾5%. The six common haplotypes in whites, found at frequencies of ⩾5%, could be distinguished using six haplotype-tagging SNPs (htSNPs) chosen from the UW-FHCRC Web site. As a secondary test, we used the PHASE package (Stephens et al. 2001) to infer haplotypes from the genotype data in the parents and children in the CAMP sample and, for confirmation, compared them with the common haplotypes seen in whites from the SeattleSNPs CEPH data. In addition to the above SNPs, we also genotyped the previously reported insertion-deletion SNP in the promoter (Morahan et al. 2002) in both the CAMP and the adult case-control studies. Figure 1 shows the location of the 6 htSNPs and the pairwise linkage disequilibrium with all other SNPs seen at ⩾5% frequency in the European CEPH sample.
Figure 1.
Pairwise linkage disequilibrium (r2) for IL12B SNPs seen at ⩾5% frequency in Europeans in CEPH sample (data from SeattleSNPs, NHLBI Program for Genomic Applications [UW-FHCRC]). SNP numbering is relative to the gene transcription start codon, with mapping to the SNPs we tested, using the SeattleSNPs numbering system.
The majority of the SNP genotyping in the CAMP cohort and in the case-control cohort was performed using the Sequenom mass spectrometry genotyping platform (Sequenom). Multiplex PCR and minisequencing assays were designed using SpectroDESIGNER software (Sequenom). Primers were purchased from Sequenom. Three–five-plex PCR reactions were performed using 5 ng of genomic DNA in 5 μl solution. Protocol details and primer data are available on the Innate Immunity in Heart, Lung and Blood Disease page of the Programs for Genomic Applications Web site. Secondary modified single-primer minisequencing reactions were formed in multiplex and were analyzed using the Bruker Bi-flex MALDI-TOF mass spectrometer (Bruker Daltonics). Spectral output was analyzed using SpectroTYPER-RT software (Sequenom). As a quality control measure, genotyping was repeated for at least 8% of the sample for each SNP and was tested for discordance.
We resequenced the promoter insertion-deletion SNP (Morahan et al. 2002) and identified the following two alleles: tctaa / −. We identified a 1-bp insertion that arose whenever there was a deletion; this was in 100% linkage disequilibrium with the insertion-deletion SNP. We used the ABI 3100 Sequence Detector (Applied Biosystems) to identify this base-pair change in both the CAMP and the adult populations.
The PedCheck program (O’Connell and Weeks 1998) was used to assess the genotype data for pedigree inconsistencies in the CAMP cohort. The genotypes of families with pedigree errors were set to 0. Hardy-Weinberg equilibrium was tested in the parental data for each locus, by use of the χ2 goodness-of-fit test.
For the CAMP cohort, the primary analysis was for association of individual SNPs with the binary trait of asthma, by use of the family-based association test (FBAT) program (Horvath et al. 2001), and for haplotypes, by use of the haplotype extension (HBAT) of the FBAT program (Horvath et al. 2004). In the FBAT program, the additive and recessive models were used. Because SNPs 4237 and 6402 were genotyped first and 2 SNP haplotypes were evaluated, we also report the 4237_6402 haplotype results.
In the CAMP cohort, we performed a second analysis, to test each individual SNP, with asthma-severity phenotypes (post-bronchodilator FEV1% predicted and baseline FEV1) and with atopy phenotypes (logarithm of total eosinophil number [LOGEOS] and logarithm of total IgE level [LOGIGE]), using PBAT (the phenotype option of FBAT) (Lange et al. 2003c). We adjusted for the following covariates, using PBAT (Lange et al. 2004), because they were statistically significant in a regression model that described the phenotypes as a function of the covariates age, age at onset, height, weight, and sex. The genetic model was selected on the basis of conditional power calculations (Lange et al. 2003a). By use of the Bonferroni correction, the significance level of the FBAT statistic was adjusted for the number of FBATs computed. Significant phenotypes were also tested for association with the SNP, by use of the population-based GEE-Wald test for family studies, as suggested by Lange et al. (2003b). The advantage of this population-based association test is that data from family-based studies can be analyzed without use of the genetic information that is included in the computation for the FBAT statistic and can therefore be considered as a replication of a significant finding in the same data set.
For the adult data set, we used linear-regression modeling for each phenotype, as a function of the marker score and other covariates. The estimates for the genetic effects and their SEs were used to construct standard Wald tests, to test for association between the SNPs and the phenotypes. By use of the technique of Pritchard and Rosenberg (1999), 49 unlinked SNPs were tested to ensure that there was no population stratification between cases and controls.
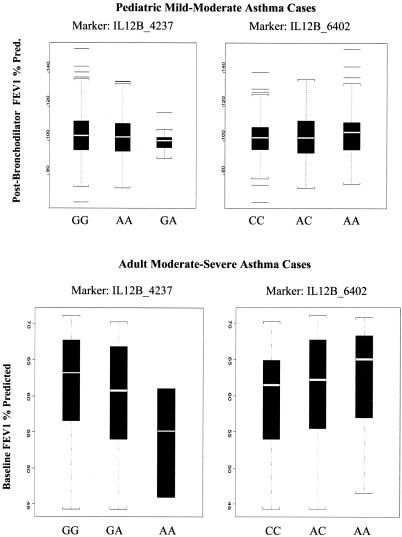
The recessive model had the most power of all three models (recessive, dominant, and additive), as evidenced by conditional power calculations and by visual inspection of the box plots (fig. 2). Therefore, we show the results using the recessive model for all analyses.
Figure 2.
Box plots of post-bronchodilator FEV1% predicted in the pediatric cohort with mild-to-moderate asthma and of baseline FEV1% predicted in the adult cases with moderate-to-severe asthma, for SNPs IL12B_4237 and IL12B_6402.
Hardy-Weinberg equilibrium was confirmed for all loci in the parents (all P values >.1). For the family-based analysis, we analyzed African Americans and whites separately. Table 2 shows the results for the recessive model for both ethnicities. The FBAT statistic is highly significant for the IL12B_4237 A allele (P=.00084) in whites, with 269 informative families. Although the FBAT statistic is significant for the G allele in African Americans (P=.0031), African Americans made up only 9.7% of our total cohort, and only 19 families were informative in this analysis. The IL12B_4237_6402 haplotype analysis was also significant for whites (global χ2 test for haplotypes 18.8; P=.0003), and the 4237A_6402C haplotype was significantly undertransmitted to asthmatic children. In addition, the analysis of all haplotypes for affection status was also significant in whites (global χ2 test for haplotypes 13.49; P=.009). By use of the recessive model, the family-based association test was also positive in whites for LOGEOS for SNP IL12B_1025 (P=.003) and for LOGIGE for SNP IL12B_4237 (P=.002), with confirmation by use of the population-based method (P=.017 and P=.05, respectively, with estimated heritabilities of 0.02 and −0.13). Because of the small number of informative families, we did not repeat the haplotype or phenotype analysis for African Americans.
Table 2.
Family-Based Association Test of IL12B SNPs in the CAMP Cohort, Using the Recessive Model
|
No. of Informative Families (P) |
||
| Markerand Allele | White | African American |
| IL12B_4237: | ||
| A | 54 (.00084) | 2 (.41421) |
| G | 215 (.38453) | 17 (.0031) |
| IL12B_13478: | ||
| C | 37 (.02304) | 3 (.7388) |
| A | 183 (.44288) | 12 (.4428) |
| IL12B_2508: | ||
| A | 127 (.9619) | 10 (.3173) |
| T | 262 (.6802) | 20 (.0495) |
In the adult asthmatic cases and nonasthmatic controls, the relative risk of asthma in cases versus controls with the IL12B_4237 SNP in a homozygous recessive model was not significant (χ2=0.1325; 1 df; P=.7158). However, the frequency of the homozygous recessives in the nonasthmatic population was only 5.7%, and the power to detect a 50% decrease in this frequency in asthmatics was only 25%. Table 3 lists the P values for all SNPs genotyped, by use of the baseline FEV1%-predicted asthma phenotype rather than the affection status, in the adult cohort, as well as the estimated heritabilities in the cohort and the FBAT statistic (recessive model) for the same SNP in CAMP, when available. The SNPs IL12B_4237 and _6402 are significantly associated with the baseline FEV1% predicted in the adult cohort. For SNPs IL12B_4237 and _6402, the associations with baseline FEV1% predicted in the adult cohort are confirmed in the CAMP study, with post-bronchodilator FEV1% predicted. Further, figure 2 shows the box plots for IL12B_4237 and _6402 in both studies. It is important to note that, for the two SNPs, the directions of the effect are the same in both studies.
Table 3.
Population-Based Analysis for Association of IL12B SNPs with Asthma Severity[Note]
|
P Value for Genotyped SNPs in |
||||
| Marker | Estimated Heritabilitya(%) | Adult Cohort | CAMP Whitesb | CAMP African Americansb |
| IL12B_4237 | 3.16 | .019 | .075 | .039 |
| IL12B_6402 | 3.40 | .014 | .016 | .098 |
Note.— Population-based analysis for association of IL12B SNPs with asthma severity, by use of the recessive model (baseline FEV1%) predicted in the adult cohort and post-bronchodilator FEV1% predicted in the CAMP cohort.
Estimated heritability is for the adult cohort and is the percentage of explained phenotypic variance.
By use of FBAT.
We subsequently tested the promoter insertion-deletion variant reported elsewhere by Morahan and colleagues (2002) to be associated with asthma severity. This SNP had r2 values for linkage disequilibrium of ⩽0.4 with the other six IL12B SNPs tested. We found no positive associations between this SNP and asthma or with any of the asthma phenotypes in either the CAMP population or the adult case-control population, using recessive, dominant, or additive models.
Our results provide strong suggestive evidence that polymorphisms in the IL12B gene are associated with asthma and asthma-related phenotypes in whites. We found, using a recessive model, that the A allele of the 4237 polymorphism was significantly undertransmitted in white asthmatic probands. In addition, the global test for IL12B haplotypes across the six SNPs was also significantly associated with asthma. We found that, in the recessive model, atopy phenotypes were associated in whites with SNPs in IL12B. We confirmed an association of IL12B with asthma severity in the CAMP cohort, in an independent adult case-control cohort, using available phenotypes (post-bronchodilator FEV1% predicted in CAMP and pre-bronchodilator FEV1% predicted in the adult population) and a recessive model. We did not find, using any model, any association of the promoter insertion-deletion SNP—reported elsewhere to be associated with asthma severity (Morahan et al. 2002)—with asthma or asthma-related phenotypes in either the CAMP or adult cohorts.
The strength of our analyses lies (1) in the replication of our findings by use of two different statistical methods (family based and population based) in the CAMP cohort and (2) in the replication of the asthma-severity finding in the CAMP cohort of mild-to-moderate asthmatics in an independent adult cohort of moderate-to-severe asthmatics. Our P values also reflect statistical controls for multiple comparisons. As a quality-control measure, we also repeated the genotyping for 50% of the CAMP sample for the two SNPs (IL12B_4237 and _6402) that were found to be associated with asthma severity. Our quality-control procedures and the use of multiple replication methods make it unlikely that genotyping error influenced our results. In addition, IL12B is a highly plausible candidate gene for asthma. It is in a location (5q31-33) previously linked repeatedly to asthma; it is associated with the development of Th1 cells, with downregulation of the release of Th2 cytokines; and it plays a major role in the mammalian innate immune response to viruses.
The limitations of our study include the fact that the two study populations used were very different. One was a cohort of children with mild-to-moderate asthma, and the other was an adult population with moderate-to-severe asthma. In the adult population, asthma-phenotypic information was limited to baseline (prebronchodilator) FEV1% predicted, and we did not replicate the exact phenotype found to be strongly associated with asthma in the adult population. This is most likely because baseline FEV1% predicted in the children with mild-to-moderate asthma was 95.1%, which is close to that of normal controls (see table 1). Other study limitations include the fact that all results were reported in the recessive model, since that was the model shown to have the most power. Because we did not measure IL12B levels, we could not correlate the genotype results with IL12B production. In addition, the case-control population was relatively small, which did not allow us to confirm the association of the IL12B_4237 SNP with the asthma phenotype in whites.
We failed to replicate the findings of Morahan and colleagues (2002) that heterozygosity for an insertion-deletion polymorphism in the IL12B promoter was associated with asthma severity. In fact, we found no association between this variant and asthma or asthma phenotypes in any population or statistical model. Khoo and colleagues (2004) also failed to find an association between asthma severity and this polymorphism in a population of asthmatic subjects for whom comprehensive data on asthma severity was followed from birth to age 42 years. This polymorphism was recently shown to be functional, with homozygotes for one allele showing a marked difference in secretion of IL12B (Muller-Berghaus et al. 2004). Although this SNP appears to be functional, our findings are consistent with those of Khoo et al. (2004), which show no association between this SNP and asthma severity.
The SNP that we found to be most positively associated with asthma was the IL12B_4237 SNP. This is an intronic SNP, and, using Neural Network Splice version 0.9 located on the Berkeley Drosophila Genome Project Web site, we found no evidence that this was a splice site. This SNP is in almost complete linkage disequilibrium with SNP IL12B_14962 (see fig. 1, where 2542=4237 and 13168=14962), an SNP in the 3′ UTR. In addition, chromosome 5q31 contains numerous immunoregulatory genes that have been linked to asthma in various studies (Meyers et al. 1994; Rosenwasser et al. 1995; Cookson and Moffatt 2000; Walley et al. 2001), and it is possible that the associations we found in IL12B are the result of linkage disequilibrium with other important SNPs in other genes.
In conclusion, we present data that suggest that multiple SNPs in IL12B are associated with asthma phenotypes in whites. Our strongest finding was for asthma-severity phenotypes. The biologic importance of these SNPs is currently unclear, and further functional testing of these SNPs—and other SNPs that are in high linkage disequilibrium with them—is required.
Acknowledgments
This work was funded by National Institutes of Health (NIH) grants K23HL04278, U01 HL66795, and PO1 HL67664 and by MedImmune Inc. A.G.R. is funded by NIH National Heart, Lung, and Blood Institute (NHLBI) K23 Award HL04278. We thank all families for their enthusiastic participation in the CAMP Genetics Ancillary Study, supported by NHLBI grant NO1-HR-16049. We also acknowledge the CAMP investigators and research team, supported by NHLBI, for collection of CAMP Genetic Ancillary Study data.
Electronic-Database Information
The URLs for data presented herein are as follows:
- Berkeley Drosophila Genome Project, http://www.fruitfly.org/
- Innate Immunity in Heart, Lung and Blood Disease, Programs for Genomic Applications, http://www.innateimmunity.net/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for IL12B) [PubMed]
- University of Washington–Fred Hutchinson Cancer Research Center (UW-FHCRC), http://pga.mbt.washington.edu/ (for SeattleSNPs, the NHLBI Program for Genomic Applications)
References
- American Thoracic Society (1987) Standards for the diagnosis and care of patients with chronic obstructive pulmonary disease (COPD) and asthma. Am Rev Respir Dis 136:225–244 [DOI] [PubMed] [Google Scholar]
- Childhood Asthma Management Program Research Group (1999) The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Control Clin Trials 20:91–120 10.1016/S0197-2456(98)00044-0 [DOI] [PubMed] [Google Scholar]
- ——— (2000) Long-term effects of budesonide or nedocromil in children with asthma. N Engl J Med 343:1054–1063 10.1056/NEJM200010123431501 [DOI] [PubMed] [Google Scholar]
- Cookson WO, Moffatt MF (2000) Genetics of asthma and allergic disease. Hum Mol Genet 9:2359–2364 10.1093/hmg/9.16.2359 [DOI] [PubMed] [Google Scholar]
- Crapo RO, Morris AH, Clayton PD, Nixon CR (1982) Lung volumes in healthy nonsmoking adults. Bull Eur Physiopathol Respir 18:419–425 [PubMed] [Google Scholar]
- Horvath S, Xu X, Laird NM (2001) The family based association test method: strategies for studying general genotype-phenotype associations. Eur J Hum Genet 9:301–306 10.1038/sj.ejhg.5200625 [DOI] [PubMed] [Google Scholar]
- Horvath S, Xu X, Lake SL, Silverman EK, Weiss ST, Laird NM (2004) Family based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol 26:61–69 10.1002/gepi.10295 [DOI] [PubMed] [Google Scholar]
- Huang D, Cancilla MR, Morahan G (2000) Complete primary structure, chromosomal localisation, and definition of polymorphisms of the gene encoding the human interleukin-12 p40 subunit. Genes Immun 1:515–520 10.1038/sj.gene.6363720 [DOI] [PubMed] [Google Scholar]
- Khoo SK, Hayden CM, Roberts M, Horak E, de Klerk N, Zhang G, Robertson CF, Goldblatt J, Le Souef P (2004) Associations of the IL12B promoter polymorphism in longitudinal data from asthmatic patients 7 to 42 years of age. J Allergy Clin Immunol 113:475–481 10.1016/j.jaci.2003.10.043 [DOI] [PubMed] [Google Scholar]
- Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM (2003a) Using the noninformative families in family-based association tests: a powerful new testing strategy. Am J Hum Genet 73:801–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (2004) PBAT: tools for family-based association studies. Am J Hum Genet 74:367–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C, Lyon H, DeMeo D, Raby BA, Silverman EK, Weiss ST (2003b) A new powerful non-parametric two-stage approach for testing multiple phenotypes in family-based association studies. Hum Hered 56:10–17 10.1159/000073728 [DOI] [PubMed] [Google Scholar]
- Lange C, Silverman EK, Xu X, Weiss ST, Laird NM (2003c) A multivariate family-based association test using generalized estimating equations: FBAT-GEE. Biostatistics 4:195–206 10.1093/biostatistics/4.2.195 [DOI] [PubMed] [Google Scholar]
- Meyers DA, Postma DS, Panhuysen CI, Xu J, Amelung PJ, Levitt RC, Bleecker ER (1994) Evidence for a locus regulating total serum IgE levels mapping to chromosome 5. Genomics 23:464–470 10.1006/geno.1994.1524 [DOI] [PubMed] [Google Scholar]
- Morahan G, Huang D, Wu M, Holt BJ, White GP, Kendall GE, Sly PD, Holt PG (2002) Association of IL12B promoter polymorphism with severity of atopic and non-atopic asthma in children. Lancet 360:455–459 10.1016/S0140-6736(02)09676-9 [DOI] [PubMed] [Google Scholar]
- Muller-Berghaus J, Kern K, Paschen A, Nguyen XD, Kluter H, Morahan G, Schadendorf D (2004) Deficient IL-12p70 secretion by dendritic cells based on IL12B promoter genotype. Genes Immun 5:431–434 10.1038/sj.gene.6364102 [DOI] [PubMed] [Google Scholar]
- Noguchi E, Yokouchi Y, Shibasaki M, Kamioka M, Yamakawa-Kobayashi K, Matsui A, Arinami T (2001) Identification of missense mutation in the IL12B gene: lack of association between IL12B polymorphisms and asthma and allergic rhinitis in the Japanese population. Genes Immun 2:401–403 10.1038/sj.gene.6363790 [DOI] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Rosenberg NA (1999) Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet 65:220–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwasser LJ, Klemm DJ, Dresback JK, Inamura H, Mascali JJ, Klinnert M, Borish L (1995) Promoter polymorphisms in the chromosome 5 gene cluster in asthma and atopy. Clin Exp Allergy Suppl 2:74–78 8590350 [DOI] [PubMed] [Google Scholar]
- Shikano H, Kato Z, Kaneko H, Watanabe M, Inoue R, Kasahara K, Takemura M, Kondo N (2001) IFN-gamma production in response to IL-18 or IL-12 stimulation by peripheral blood mononuclear cells of atopic patients. Clin Exp Allergy 31:1263–1270 10.1046/j.1365-2222.2001.01141.x [DOI] [PubMed] [Google Scholar]
- Sonna LA, Zhao L, Angel KC, Cullivan M, Lilly C, US Army Research Institute of Environmental Medicine (USARIEM) (2000) Environmental Medicine Genome Bank Current Composition. TN 0-8 (available at http://www.stormingmedia.us/08/0810/A081083.html?searchTerms=~sonna,~zhao,~angel [accessed August 13, 2004])
- Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinchieri G, Scott P (1994) The role of interleukin 12 in the immune response, disease and therapy. Immunol Today 15:460–463 10.1016/0167-5699(94)90189-9 [DOI] [PubMed] [Google Scholar]
- Walley AJ, Wiltshire S, Ellis CM, Cookson WO (2001) Linkage and allelic association of chromosome 5 cytokine cluster genetic markers with atopy and asthma associated traits. Genomics 72:15–20 10.1006/geno.2000.6435 [DOI] [PubMed] [Google Scholar]
- Yap G, Pesin M, Sher A (2000) Cutting edge: IL-12 is required for the maintenance of IFN-gamma production in T cells mediating chronic resistance to the intracellular pathogen, Toxoplasma gondii. J Immunol 165:628–631 [DOI] [PubMed] [Google Scholar]