Figure 3.
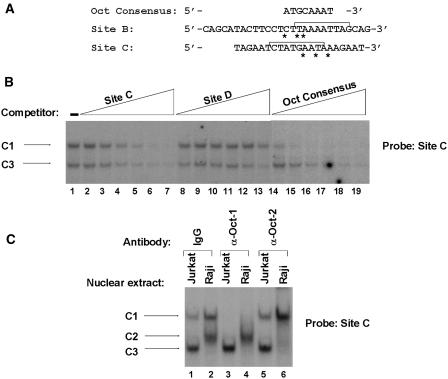
Oct-1 and Oct-2 specifically interact with site C. (A) The nucleotide sequence of the wild-type sites B and C oligonucleotides are aligned with the octamer consensus motif. The asterisks indicate the mismatches of the potential site B and site C octamer motifs with respect to the octamer consensus. The brackets in site B and site C show the 9 bp (identified in Figure 2) as critical for the formation of complexes B1, B2 and B3 and C1, C2, and C3, respectively. (B) The site C oligonucleotide probe was incubated with nuclear extracts from Jurkat cells. Binding assays were performed in the absence of competitor (lane 1) or in the presence of increasing concentrations (5-, 10-, 25-, 50-, 100- or 200-fold molar excess) of the homologous site C oligonucleotide (lanes 2–7), of the heterologous HS7 site D oligonucleotide (lanes 8–13) or of the Oct consensus oligonucleotide (lanes 14–19). The sequence of the coding strand of the Oct consensus oligonucleotide was as follows: 5′-TGTCGAATGCAAATCACTAGAA-3′. The sequence of the coding strand of the HS7 site D oligonucleotide is shown in Figure 7. The figure shows only the specific retarded bands of interest. The complexes C1 and C3 are indicated by arrows. (C) Antibodies directed against Oct-1 (lanes 3 and 4) and Oct-2 (lanes 5 and 6) or purified rabbit IgG as negative control (lanes 1 and 2) were incubated with 15 μg of nuclear extracts from Jurkat (lanes 1, 3 and 5) or Raji (lanes 2, 4 and 6) cells before the addition of the site C oligonucleotide probe. The figure shows only the specific retarded bands of interest. The retarded DNA–protein complexes C1, C2 and C3 are indicated by arrows.