Abstract
An alternative technology for the estimation of T cells based on a microcapillary technique (Guava Technologies, Hayward, CA) was compared to FACSCount (Becton Dickinson, San Jose, CA). Samples from 51 human immunodeficiency virus-infected and 21 healthy individuals were tested. The correlation (r) of the two systems for CD4+ T cells was 0.994, and the coefficient of variation was 6.5%, establishing equable performance between the two technologies.
The number of human immunodeficiency virus (HIV)-infected individuals continues to rise in India ever since it was first detected in Vellore, India (6). A high proportion of these infected individuals belong to the economically lower strata of society, and hence it is essential that the supporting health care systems operate in a cost-effective manner. CD4+ T-cell counts are essential for monitoring disease progression and as indicators of when treatment should be started (4). Flow cytometry is the widely used method for analyzing CD4 counts, including in India. More recently a system based on a new technology developed by Guava Technologies (Hayward, CA) has been introduced. This technology has been used only in a small number of centers and requires more widespread testing for user acceptability. In this study, we investigated the performance of the Guava EasyCD4 System in relation to FACSCount.
Blood samples were collected between 8:00 a.m. and 10:00 a.m. from 51 HIV-infected individuals who had come to the Clinical Virology department of a tertiary care center in India (south) for their CD4+/CD8+ T-cell estimation and also from 21 normal healthy individuals from October 2004 through December 2004 after informed consent was obtained in K2 EDTA vacutainer tubes. Of the 51 samples from HIV-infected individuals, 8 were done in duplicates to investigate the reproducibility of results. In addition to these eight samples, samples from four HIV-infected individuals were tested in quadriplicates. Samples from another seven HIV-infected individuals were stored at room temperature (20 to 25°C) and were retested after 24 h to look for any difference in the count. In order to examine the interpersonnel variation in testing, samples from two healthy individuals were processed and tested by five different laboratory personnel only for CD4 counts.
Absolute CD4+/CD8+ T-cell counts were estimated by the Guava EasyCD4 System, which allows enumeration of CD4+ and/or CD8+ lymphocytes in human peripheral blood. The instrument used two fluorescence parameters in combination with forward scatter (FSC) to identify cells. The reagents consisted of a monoclonal anti-human CD3 antibody conjugated to the tandem dye phycoerythrin (PE)-Cy5 and monoclonal anti-human CD4 and CD8 antibodies conjugated to PE. The antibody staining solutions were used to stain 10-μl portions of the blood samples. For each sample, two separate tubes were used: one for determining the absolute CD4+ T-cell count (monoclonal antibody CD3PE-Cy5 and CD4PE were added) and the other for determining the CD8+ T-cell count (monoclonal antibody CD3PE-Cy5 and CD8PE were added). The 1× lysing solution was then added after a 15-min incubation period at room temperature (20 to 25°C). After another 15 min of incubation at room temperature, the samples were screened, and the data were acquired on the Guava Personal Cell Analysis (PCA) instrument by using CytoSoft version 2.2 software.
The comparison of the above technique was carried out by parallel testing of 72 samples using FACSCount (Becton Dickinson Immunocytometry Systems, San Jose, CA) and 66 samples by Guava EasyCD4 system (Guava Technologies, Hayward, CA) at the YRG CARE Infectious Diseases Laboratory, Chennai, India. CD4+/CD8+ T-cell enumeration was done for each specimen with a two-color single-platform flow cytometer, FACSCount, according to the manufacturer's instructions. The testing at laboratory 1 (CMC) was done within 5 h of sample collection, and the testing at the second laboratory at Chennai was done within 10 h of collection.
The correlation between the techniques was assessed by the Pearson correlation test. The percent coefficient of variation (%CV) for the mean cell counts estimated by the two different techniques and the Guava EasyCD4 counts estimated at two different centers were calculated by using Microsoft Excel software. We also analyzed the data using Bland-Altman plots by displaying the average values of cells counts obtained by both methods on the x axis and the difference between the two methods shown on the y axis (2).
For purpose of analysis, all values estimated by FACSCount indicated as <50 cells/μl was considered as 49 cells/μl, and all values of >2,000 cells/μl was taken as 2,001 cells/μl. Based on the CD4+ T-cell counts obtained with the FACSCount, the HIV-infected individuals were classified into three CDC categories (3). There were 7 individuals in category 1, 24 individuals in category 2, and 20 individuals in category 3.
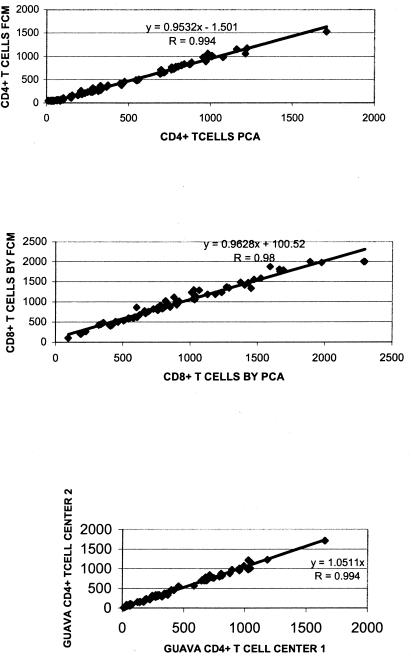
The mean, median, and 10th and 90th percentile differences for the absolute CD4+ and CD8+ T cells, as determined by FACSCount and Guava EasyCD4 system, among the healthy individuals, the three CDC categories of HIV-infected patients, and all 72 study volunteers are shown in Table 1. A scattergram showing the correlation for both CD4+ T cells and CD8+ T cells measured by both the techniques and the correlation between the CD4+ T cells estimated by the Guava easyCD4 system at two different centers are shown in Fig. 1.
TABLE 1.
Mean, median, and 10th and 90th percentile differences for absolute CD4+ and CD8+ T cells as determined by FACSCount and Guava EasyCD4 methods among control subjects and three CDC categories of HIV-infected patients
| Group | n | Mean, median, or percentile | Value determined for the indicated cells by:
|
|||
|---|---|---|---|---|---|---|
| FACSCount
|
Guava EasyCD4
|
|||||
| CD4+ cells/μl | CD8+ cells/μl | CD4+ cells/μl | CD8+ cells/μl | |||
| Healthy individuals | 21 | Mean | 913 | 844 | 946 | 758 |
| 10th Percentile | 716 | 529 | 760 | 507 | ||
| 90th Percentile | 1,149 | 1,235 | 1,215 | 1,042 | ||
| Median | 866 | 827 | 884 | 689 | ||
| HIV-infected individuals | ||||||
| CDC category 1 | 7 | Mean | 774 | 1,553 | 819 | 1,545 |
| 10th Percentile | 576 | 1,030 | 640 | 981 | ||
| 90th Percentile | 989 | 2,001 | 1,036 | 2,100 | ||
| Median | 803 | 1,489 | 824 | 1,405 | ||
| CDC category 2 | 24 | Mean | 320 | 1,174 | 347 | 1,118 |
| 10th Percentile | 232 | 584 | 258 | 546 | ||
| 90th Percentile | 446 | 1,803 | 471 | 1,666 | ||
| Median | 287 | 1,086 | 324 | 966 | ||
| CDC category 3 | 20 | Mean | 86 | 682 | 92 | 603 |
| 10th Percentile | 49 | 204 | 30 | 190 | ||
| 90th Percentile | 164 | 1,200 | 162 | 1,026 | ||
| Median | 63 | 605 | 77 | 564 | ||
| All study volunteers | 72 | Mean | 472 | 978 | 497 | 911 |
| 10th Percentile | 57 | 429 | 62 | 403 | ||
| 90th Percentile | 977 | 1,743 | 984 | 1,586 | ||
| Median | 352 | 918 | 350 | 819 | ||
FIG. 1.
Correlation and r values for CD4+ (top) and CD8+ (middle) T-cell counts estimated by Guava EasyCD4 (PCA) with FACSCount (FCM) and the CD4+ T-cell counts (bottom) estimated by Guava EasyCD4 (PCA) at two centers.
The mean %CV calculated for the CD4+ T-cell counts (excluding six samples that gave <50 CD4+ T cells by FACSCount) between the Guava EasyCD4 System and FACScount was 6.5% and that for CD8+ T cells (excluding three samples that gave >2,000 CD8+ T cells by FACSCount) was 6.67%. The mean %CV calculated for the CD4+ T-cell counts estimated by the Guava EasyCD4 system at two different centers (n = 66) was 5.39%.
The mean coefficients of variation between the cell counts estimated (n = 6) on the same day and after 24 h were 2.62 (standard deviation [SD] = 2.51) and 1.74 (SD = 1.23), respectively, for CD4+ and CD8+ T cells. Duplicate testing of eight samples showed mean coefficients of variation of 3.44 (SD = 2.84) and 3.65 (SD = 2.62) for CD4+ and CD8+ T cells, respectively. In addition, four samples were tested in quadruplicate; the mean coefficients of variation were 6.8 (SD = 2.50) and 3.9 (SD = 1.59) for CD4+ and CD8+ T cells, respectively. One of the two samples tested by five different individuals had a mean of 596.60 cells/μl (SD = 1.67), and for the other sample the mean was 1,200 (SD = 85).
As antiretroviral treatment becomes cheaper and more accessible in developing countries, HIV-infected individuals in countries such as India need an affordable and reliable system for monitoring their immune status by the estimation of T-cell subsets. Our evaluation of the Guava EasyCD4 System, a microcapillary cytometry for the estimation of CD4+/CD8+ T cells shows it to be an economically viable and a reliable alternative for CD4+/CD8+ T-cell estimation. The overall correlation of the Guava EasyCD4 System to the FACSCount for CD4+ T cells was 0.994, and for CD8+ T cells it was 0.98.
The reported interlaboratory %CV for “single-platform” systems is about 13.7% (10 to 18.3%). However, the %CV reported for “double-platform” systems are comparatively high, ranging form 14.5 to 43.4% (mean, 23.4%) (1). This %CV observed in our study between the Guava EasyCD4 System and FACSCount in two different laboratories was only 6.5%. The %CV observed for the Guava EasyCD4 System at two different centers was even less (5.39%). The %CV valuess observed in our study were thus even lower than the minimum (10%) reported interlaboratory variation for the single-platform system.
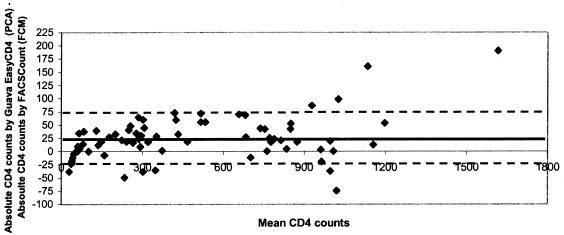
The Bland-Altman plot analysis of our data showed that the two methods agree sufficiently and can be used interchangeably. The CD4+ T-cell counts estimated by the Guava EasyCD4 System was slightly higher at our center (bias, +25; 95% confidence interval [CI] +15 to +34.3, limits of agreement between +75 and −25) than the FACSCount (Fig. 2). However, slightly lower CD8+ T-cell counts were observed on Guava PCA (bias, -66.6; 95% CI, −47 to −88; limits of agreement between +54 and −188). The 66 samples tested by the Guava EasyCD4 System at the second center yielded a slightly lower CD4 count (bias, -6; limits of agreement between +76 and −65). Of the 20 individuals who belonged to CDC category 3 as determined by FACSCount, the CD4 T-cell counts were <200 as determined by the Guava EasyCD4 System in 19 individuals, and 1 individual had a cell count of 217. Similarly, among the 24 CDC category 2 individuals as determined by FACSCount, only 2 showed a CD4+ T-cell count of >500. These differences in cell counts observed between assays and laboratories were well with in the inter laboratory cell count reported for a single sample (5). Hence, these two systems can be used interchangeably.
FIG. 2.
Bland-Altman plot comparing absolute CD4 cell counts estimated by Guava EasyCD4 (PCA) and FACSCount (FCM). The dark continuous line drawn indicates the bias (mean difference), and the dotted lines are the limits of agreement (mean ± 2 SD). Mean, +25; limits of agreement between +75 and −25.
The mean coefficients of variation between the cell counts estimated on the day of sample collection and the following day were 2.62 (SD = 2.51) and 1.74 (SD = 1.23), respectively, for CD4+ and CD8+ T cells. This lack of change in cell counts after 24 h of testing is of advantage in resource-poor countries when field areas are not in close proximity to laboratories and where there is a likelihood of delay in transportation of samples.
The FACSCount from Becton Dickinson (San Jose, CA) and the Guava PCA from Guava Technologies (Hayward, CA) are sold in the Indian market at comparable prices of ca. $47,000 (U.S. dollars). The BD reagents are slightly more expensive than that for Guava PCA. CD4/CD8 estimation by FACSCount is approximately $18 per test, and that by Guava PCA is approximately $10, excluding the overheads, capital depreciation, and labor costs.
The Guava EasyCD4 System is a user-friendly technique in which the involvement of different laboratory personnel performing this assay are unlikely to cause any significant variation in the cell count values, as shown in our study.
In summary, we find the Guava EasyCD4 System is a reliable and valid alternative for CD4+ T-lymphocyte estimation in HIV-infected individuals. This study establishes equable performance between the new (Guava EasyCD4) System and the more widely used conventional technology (FACSCount).
Acknowledgments
No conflict of interest exists since the laboratories of the authors had purchased the respective equipment and reagents.
REFERENCES
- 1.Barnett, D., V. Granger, L. Whitby, I. Storie, and J. T. Reilly. 1999. Absolute CD4+ T-lymphocyte and CD34+ stem cell counts by single-platform flow cytometry: the way forward. Br. J. Haematol. 106:1059-1062. [DOI] [PubMed] [Google Scholar]
- 2.Bland, J. M., and D. G. Altman. 1986. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet i:307-310. [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 1992. 1993 revised classification system for HIV infection and expanded surveillance case definition for AIDS among adolescents and adults. Morb. Mortal. Wkly. Rep. Recomm Rep. 41(RR-17):1-19. [PubMed] [Google Scholar]
- 4.Kaplan, J. E., H. Masur, and K. K. Holmes. 2002. Guidelines for preventing opportunistic infections among HIV-infected persons-2002: recommendations of the U.S. Public Health Service and the Infectious Diseases Society of America. Morb. Mortal. Wkly. Rep. Recomm. Rep. 51(RR-8):1-52. [PubMed] [Google Scholar]
- 5.Kunkl, A., D. Risso, M. P. Terranova, M. Girotto, B. Brando, L. Mortara, and P. B. Lantieri. 2002. Grading of laboratories on CD4+ T-lymphocyte evaluations based on acceptable data boundaries defined by the measurement error. Cytometry 50:117-126. [DOI] [PubMed] [Google Scholar]
- 6.Simoes, E. A. F., P. G. Babu, T. J. John, S. Nirmala, S. Solomon, C. S. Lakshminarayana, and T. C Quinn. 1987. Evidence for HTLV-III infection in prostitutes in Tamil Nadu (India). Ind. J. Med. Res. 85:335-338. [PubMed] [Google Scholar]