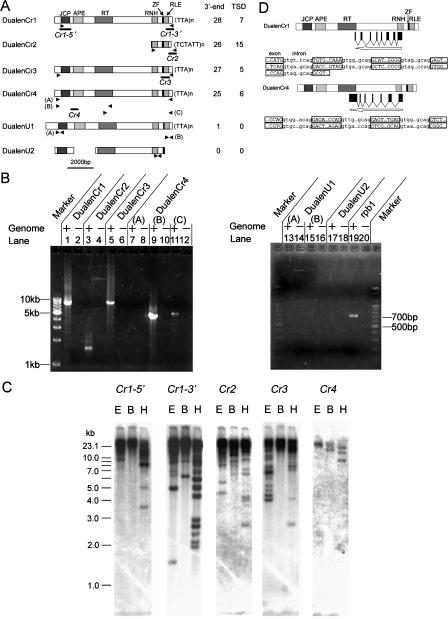
Figure 1.
Characterization of Dualen. (A) Structure of six Dualen elements. Motifs and domains are schematically shown as boxes. The numbers of identified 3′-ends and target site duplications (TSDs) are listed at right. Primers used for PCR (B) are shown as arrowheads, and regions used for probes in Southern hybridization (C) are shown as bold lines below the structure. (JCP) Josephin-related cysteine protease; (APE) apurinic/apyrimidinic endonuclease-like endonuclease; (RT) reverse transcriptase; (RNH) ribonuclease H; (ZF) zinc-finger motif; (RLE) restriction-like endonuclease. (B) Confirmation of the existence of Dualen elements by PCR. Primers used for PCR and expected sizes of PCR products are as follows: (lanes 1,2) DualenCr1_F2 and DualenCr1_R1, 8740 bp; (lanes 3,4) DualenCr2_F2 and DualenCr2_R1, 1659 bp; (lanes 5,6) DualenCr3longF1 and DualenCr3longR1, 9233 bp; (lanes 7,8) DualenCr4longF1 and DualenCr4longR1, 10,075 bp; (lanes 9,10) DualenCr4longF1 and DualenCr4longR3, 4940 bp; (lanes 11,12) DualenCr4longF3 and DualenCr4longR1, 5701 bp; (lanes 13,14) DualenAt1_F1 and DualenAt1_R1, 610 bp; (lanes 15,16) DualenAt1_F2 and DualenAt2_R2, 462 bp; (lanes 17,18) DualenAt2_F and DualenAt2_R, 467 bp; (lanes 19,20) Atrpb1F and Atrpb1R, 707 bp. Primers used for PCR are listed in Supplemental Table 1. (C) Southern hybridization. Probes are shown in A. (E) EcoRI; (B) BglII; (H) HindIII. (D) Diagrams of spliced antisense RNAs. Exon–intron boundaries of antisense RNAs are shown below. Exon sequences are in uppercase and boxed, while intron sequences are in lowercase.