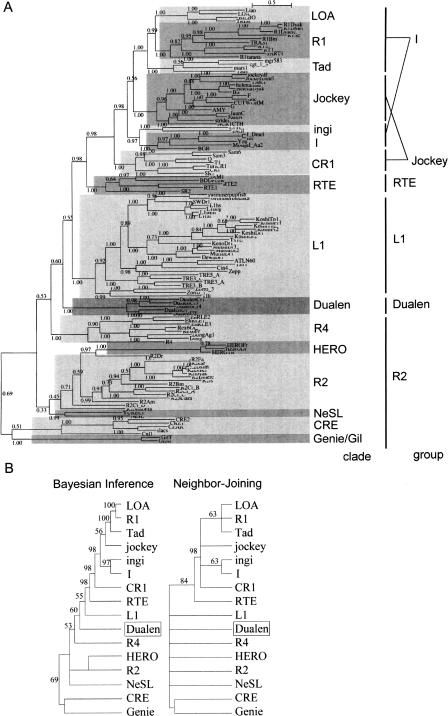
Figure 2.
Phylogenetic analyses of reverse transcriptase (RT). (A) The Bayesian phylogenetic inference. Posterior probabilities are indicated. Markov chain Monte Carlo (MCMC) chain length was 500,000 generations with trees sampled every 10 generations; the first 3300 trees were discarded as burn-in. (B) The 50% consensus trees of the Bayesian inference and the Neighbor-Joining methods. Retrotransposons belonging to the same clade are compressed. Posterior probabilities (Bayesian inference) and bootstrap values (Neighbor-Joining) are indicated as a percentage. HERO is not recognized as clade, but treated independently here due to its indefinite position. The sources of sequences in the L1, R4, HERO, R2, NeSL, CRE, and Genie/Gil clades are given in our previous study (Kojima and Fujiwara 2004). Others are given in the report by Malik et al. (1999). The alignment used to generate this tree is based on two previous alignments, ds36752 (Malik et al. 1999) and ALIGN 00231 (Burke et al. 2002), and available as Supplemental Figure 1.