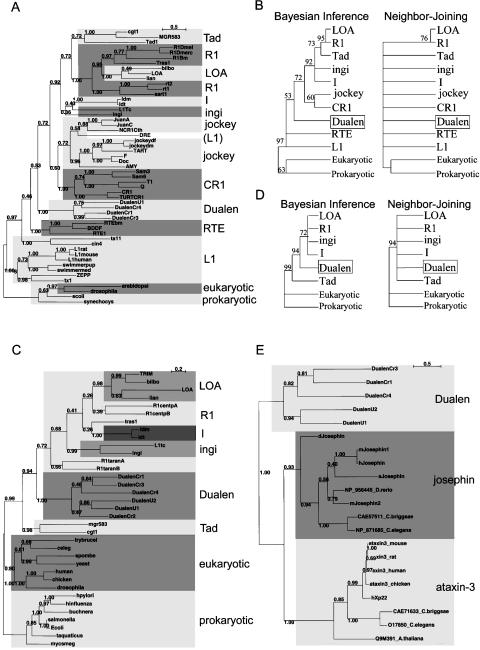
Figure 5.
Phylogenetic analyses of apurinic/apyrimidinic endonuclease-like endonuclease (APE), ribonuclease H (RNH), and josephin-related cysteine protease (JCP). (A) The Bayesian phylogenetic inference tree of APE. Posterior probabilities are indicated. MCMC chain length was 150,000 generations with trees sampled every 10 generations; the first 1000 trees were discarded as burn-in. (B) The 50% consensus APE trees of the Bayesian inference and the Neighbor-Joining methods. Retrotransposons belonging to the same clade are compressed. Posterior probabilities (Bayesian inference) and bootstrap values (Neighbor-Joining) are indicated as a percentage. (C) The Bayesian phylogenetic inference of RNH. MCMC chain length was 100,000 generations with trees sampled every 10 generations; the first 700 trees were discarded as burn-in. (D) The 50% consensus RNH trees of the Bayesian inference and the Neighbor-Joining methods. (E) The Bayesian phylogenetic inference of JCP and josephin. MCMC chain length was 100,000 generations with trees sampled every 10 generations; the first 400 trees were discarded as burn-in.