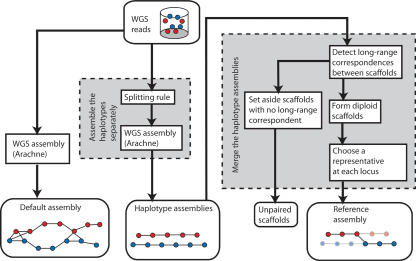
Figure 3.
Our method for diploid genome assembly. The multistep process has two main components, which were motivated by an analysis of the default assembly using Arachne. First, we assemble the two haplotypes separately by applying a new algorithm called “the splitting rule” prior to the formation of contigs and scaffolds. This forces the two haplotypes to assemble separately. Second, we merge the two haplotypes by detecting long-range correspondences between haploid scaffolds, forming diploid scaffolds, and then choosing the best representative at each locus. When we cannot unambiguously determine the partner of a haploid scaffold, we set it aside as an unpaired scaffold.