Figure 4.
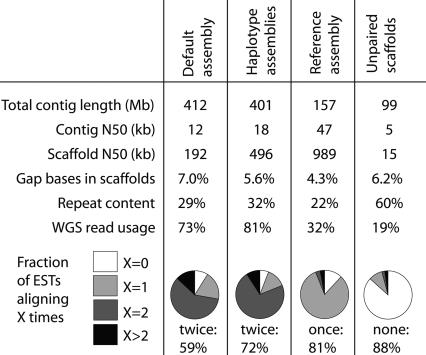
The results of applying our method for diploid genome assembly to the ∼13× WGS data set for Ciona savignyi. The majority of ESTs align exactly twice to the default assembly using Arachne, indicating a tendency for the two haplotypes to assemble separately. The fraction of ESTs aligning twice has increased in the haplotype assemblies, consistent with the goal of separating the haplotypes more cleanly. The process of merging the haplotype assemblies had three effects: representing the vast majority of loci exactly once instead of twice, increasing the scaffold size, and increasing the contig size. We also report the unpaired scaffolds, for which we were unable to determine their partner of the opposite haplotype—on average they are small, highly repetitive, and depleted for EST alignments.