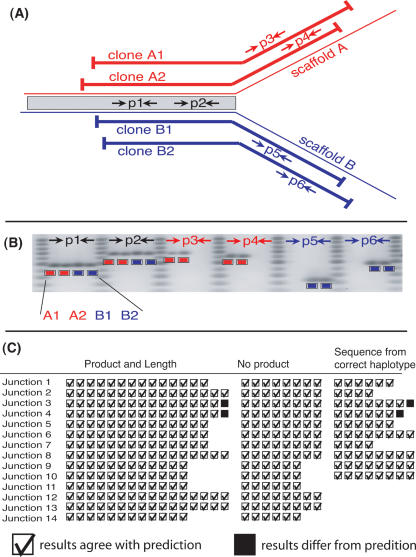
Figure 5.
Experimental design for validating the assembly at critical junctions, sample results, and summary of results from 14 critical junctions. At a critical junction (at which the haploid scaffolds correspond up to a point and then cease to correspond) we select two 40-kb clones from each haplotype spanning the junction. We check that the draft sequences correspond to the clone sequences by designing six PCR assays that are applied to each clone (A). For these 24 assays, we predict a product and its length in 16 assays and the absence of a product in eight assays, as in the sample results (B). We also sequenced the PCR products at 10 of the critical junctions and checked that they align best to the draft sequence of the correct haplotype. Accounting for variations in experimental design, we made predictions for 304 PCR assays (see Supplemental material). We observed only two discrepant PCR product lengths, and the nucleotide sequence of these two products was also discrepant (C). The overwhelming agreement between prediction and observation supports the interpretation that the large-scale haplotype differences are real and that we assembled them correctly.