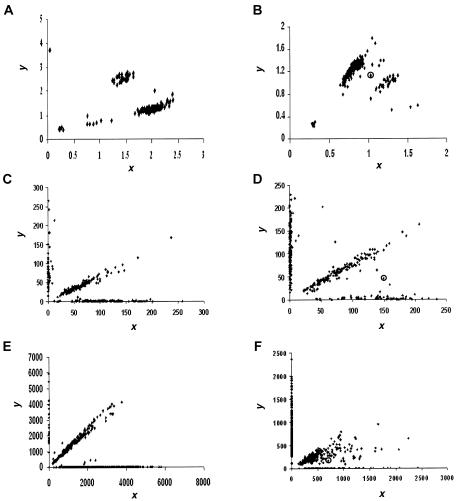
Figure 1.
Scatterplots of FI readouts from genotyping a marker by use of various assays. Each point (x, y) represents the genotype of an individual, where x and y denote the FI values for the two alleles, respectively. A, A typical good result from the TaqMan assay. Four distinct clusters are shown, corresponding to major-allele homozygotes, minor-allele homozygotes, heterozygotes, and NFS. B, A typical but not ideal result from the TaqMan assay. It is difficult to separate all points into distinct clusters. The point in a circle is located between two groups of dense points, demonstrating the case in which a clear-cut genotype call is difficult to make. C, A typical good result from the OLA. The three genotype clusters are in the form of three straight lines: the one close to the x-axis and the one close to the y-axis correspond to major and minor homozygotes respectively, and the center line corresponds to heterozygotes. The points near the origin indicate experimental failures, resulting in NFS. D, A typical but not ideal result from the OLA. The points located between line patterns demonstrate the cases in which a clear-cut genotype call is difficult to make. E, A typical good result from the MassARRAY assay. The scatterplot looks similar to the ones obtained from the OLA. F, A typical but not ideal result from the MassARRAY assay. The points that are located between the genotype line patterns are the cases in which a clear-cut genotype call is difficult to make.