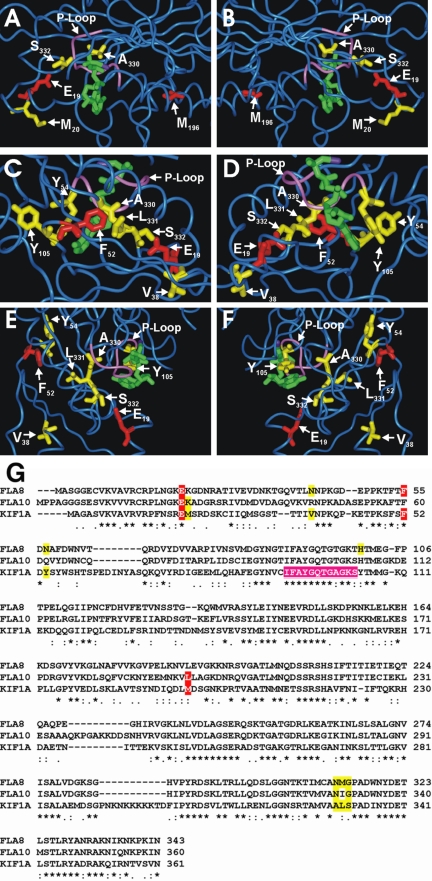
Figure 5.
Location of amino acids altered in the fla8 and fla10 mutant and revertant alleles as modeled onto the crystal structure of KIF1A with AMPPNP (green) bound (PDB 1VFV). The P-loop is shown in purple. (A and B) Fla10 E24 and L196 correspond to KIF1A E19 and M196, respectively, and are shown in red. Revertant fla10-14 amino acids K25, N329, and G331 correspond to KIF1A M20, A330, and S332, respectively, and are shown in yellow. (C–F) Fla8 amino acids E21 and F55 correspond to KIF1A E19 and F52, respectively, and are shown in red. Revertant fla8-1 and fla8-2 amino acids N42, N57, H100, N312, M313, and G314 correspond to KIF1A V38, Y54, Y105, A330, L331, and S332, respectively, and are shown in yellow. (G) Alignment of Fla8, Fla10, and KIF1A in the regions of interest. Red shading indicates mutations in one of the kinesin-2 motor subunits, yellow shading indicates amino acids that are altered in the revertants, and purple shading indicates the P-loop residues.