Abstract
The maturation of 5S ribosomal RNA in low G+C Gram-positive bacteria is catalyzed by a highly conserved, ∼190 residue, enzyme, called ribonuclease M5 (RNase M5). Sequence alignment had predicted that the N-terminal half of RNase M5 would consist of a Toprim domain, a protein fold found in type IA and type II topoisomerases, DnaG-like primases, OLD family nucleases and RecR proteins [L. Aravind, D. D. Leipe and E. V. Koonin (1998) Nucleic Acids Res., 26, 4205–4213]. Here, we present structural modelling data and a mutational analysis of RNase M5 that confirms this hypothesis. The N-terminal half of RNase M5 can be fitted to the Toprim domain of the DnaG catalytic core. Mutation of amino acid residues highly conserved among RNase M5 enzymes and members of the Toprim domain family showed that alteration of residues critical for topoisomerase and primase activity also had a dramatic effect on the cleavage of 5S rRNA precursor by RNase M5 both in vivo and in vitro. This suggests that the mechanisms of double-stranded RNA cleavage by RNase M5 and double-stranded DNA cleavage by members of the topoisomerase family are related.
INTRODUCTION
In Bacillus subtilis, and in other low G+C containing Gram-positive bacteria, 5S ribosomal RNA processing is performed by an enzyme known as ribonuclease M5 (RNase M5) (1,2). In the presence of ribosomal protein L18, RNase M5 cleaves 5S rRNA precursor on both sides of a double-stranded stem, to yield mature 5S rRNA in one step (3). L18 binds 5S rRNA and has been proposed to act as an RNA chaperone by allowing the 5S rRNA precursor to adopt the correct conformation for cleavage, rather than playing a direct role in the processing reaction itself (4). Ribosomal protein L5, which also binds 5S rRNA, inhibits the cleavage reaction at high concentrations (3). RNase M5 is encoded by the rnmV gene, which is not essential for B.subtilis viability, despite the fact that strains lacking a functional copy of this gene produce no mature 5S rRNA whatsoever (2). Instead, 5S rRNA is found as precursor species in both ribosomes and polysomes, differing in length depending on from which of the 10 ribosomal RNA operons (rrn) they originated. The ΔrnmV mutation has no major effect on growth rate; thus, 5S rRNA maturation is not essential for efficient ribosome function in B.subtilis. RNase M5 appears to have no other function in the cell apart from 5S rRNA maturation (5).
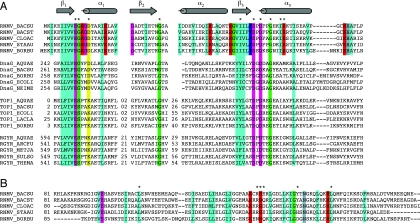
The B.subtilis RNase M5 protein (previously known as YabF) was predicted to contain a Toprim domain at its N-terminal end by sequence and secondary structure alignment (6). This ancient domain is found in type IA and type II topoisomerases, DnaG-type primases, nucleases of the P2 phage OLD family and RecR proteins. The crystal structures of four proteins containing a Toprim domain are known, those of Escherichia coli topoisomerase I (TopI) and DnaG primase, an archaeal reverse gyrase (r-gyrase) from Archeoglobus fulgidus and Deinococcus radiodurans RecR (7–10). This domain is characterized by a compact βαβαβαβ fold, with the four hydrophobic β-strands forming a central core, bounded on three sides by α-helices. The Toprim domain is also defined by three motifs, a glutamate residue in the loop between β1 and α1, a glycine residue in the loop between β2 and α2 and a DxDxxG sequence, comprising the beginning of helix α3 and the preceding loop (Figure 1).
Figure 1.
The Toprim domain. (A) Alignment of the Toprim domains of RNase M5, DnaG primase, Topoisomerase I and reverse gyrase from different bacteria. BACSU, Bacillus subtilis; BACST, Bacillus stearothermophilus; CLOAC, Clostridium acetobutylicum; STAAU, Staphylococcus aureus; BORBU, Borrelia burgdorferi; AQUAE, Aquifex aeolicus; ECOLI, Escherichia coli; NEIME, Neisseria meningitidis; LACLA, Lactococcus lactis; ARCFU, Archeoglobus fulgidus; METJA, Methanococcus jannaschii; SULSO, Sulfolobus sulfataricus; THEMA, Thermotoga maritima. The colour codes are as follows: turquoise for hydrophobic (A, C, F, I, L, M, V, W, Y), mauve for negatively charged (E, D), green for small (G, S, A), yellow for charged (E, D, R, K, H) and red for positively charged (R, K). Conserved amino acids chosen for mutagenesis are marked with an asterisk. (B) Alignment of the C-terminal domain of RNase M5. Colour codes are the same as in part (A).
The similarity to the topoisomerases is of particular interest, as these enzymes cleave double-stranded DNA to permit strand passage in the process of altering the extent and nature of DNA supercoils. Furthermore, two groups have shown that type I topoisomerases can cleave RNA under certain circumstances (11,12). The mechanism of DNA and RNA cleavage by type I topoisomerases is known. The hydroxyl group of an active site tyrosine residue attacks the scissile phosphodiester bond producing a covalently linked protein–RNA intermediate. This reaction is essentially reversed following strand passage to allow religation of the substrate. The DxD motif of the Toprim domain is involved in the co-ordination of magnesium ion, similar to its role in the Klenow fragment of DNA polymerase I (13,14). Indeed, all members of the Toprim family require Mg2+ for activity. The conserved glutamate has been proposed to play a dual role as a general acid for cleavage, by donating a proton to the sugar hydroxyl, and as a general base for religation, by abstracting it (15). It may also be involved in Mg2+ co-ordination (16).
In this study, we have probed the structural relationship between the proposed Toprim domain of RNase M5 and other members of this family. We show that we can thread the N-terminal half of RNase M5 onto the Toprim domain structure of the catalytic core of E.coli DnaG primase. We also show that amino acids important for topoisomerase and primase cleavage activity are also critical for RNase M5 activity both in vivo and in vitro. These data confirm that RNase M5 is related to the ancient Toprim domain family of enzymes involved in DNA replication and repair.
MATERIALS AND METHODS
Plasmid construction and mutagenesis
All oligonucleotides used in this study are given in Supplementary Table I.
The B.subtilis rnmV gene was cloned as a PCR-amplified fragment between the EcoRI and HindIII sites of pBluescript KS+. The oligonucleotides used for amplification, HP419 and HP420, had integrated restriction sites and an in-frame hexahistidine coding C-terminal tag. The resulting plasmid, pHMM9, was confirmed by sequencing (MWG). Mutations were introduced into the rnmV gene of pHMM9 by the Kunkel (17) or QuikChange™ site-directed mutagenesis methods (Stratagene). Plasmids were named pHMM9-353, pHMM9-359, etc. according to the name of the oligo used to introduce the mutation. Mutants were confirmed by sequencing, and then transformed into E.coli strain BL21 CodonPlus (Stratagene) for overexpression by T7 RNA polymerase. The Y152A mutant was subcloned in pET28a (Novagen) for overproduction and purification.
Modelling of the RNase M5 Toprim domain
We searched for potential structural homologues of RNase M5 using PSI-BLAST (18) against the non-redundant database (BLOSUM 62 matrix). After the sixth iteration, we found an E-value of 7 × 10−33 and 23% identity with the catalytic core of E.coli DNA primase, DnaG (PDB entry 1DD9) (8). This is the well-known fold known as the Toprim domain (6).
The secondary structure of RNase M5, predicted by the PSIPRED program (19), was compared with the secondary structure of DnaG, produced by PDBsum. Hydrophobic Cluster Analysis (HCA) was also used to fine-tune the secondary structure alignment (20,21).
We then compared the RNase M5 sequence with the 3D amino acid profiles of proteins with known structures. For this, we used the CBS-Metaserver (22), which included the 3D-PSSM (23), GenTHREADER (24), FUGUE (25), SAM-T99 and JPRED2 methods. A significant score was obtained for the 1DD9 structure by the 3D-PSSM program.
The compatibility of the amino acid sequence of RNase M5 and the 3D structure of DnaG Toprim domain was evaluated by TITO (Tool for Incrementation Threading Optimisation), used for low identity (<20%) scores (26). TITO alignment was manually modified to respect the HCA profile and submitted to Modeller 3D (27) on the CBS-Metaserver (http://bioserv.cbs.cnrs.fr/HTML_BIO/frame_meta.html). The model proposed by Modeller 3D was validated by PROSAII (28). The resulting domain could be superimposed on the Toprim domain of the DnaG structure with a root-mean-square deviation of 4.8 Å between the α-carbons of Leu6–Pro81 of RNase M5 and Leu261–Phe337 of DnaG.
Introduction of rnmV mutants on the B.subtilis chromosome
The rnmV gene of B.subtilis strain W168 was first interrupted by an erythromycin resistance antibiotic cassette (ery), by transformation with pBS-M5-Ery, creating strain CCB038 (ΔrnmV::ery). Plasmid pBS-M5-Ery consists of the ery cassette, bounded by ∼500 bp regions upstream and downstream of rnmV, cloned in pBluescript KS+. The upstream and downstream fragments were amplified from B.subtilis W168 chromosomal DNA using the oligo pairs CC005/CC006 and CC007/CC008, respectively. The ery cassette was amplified from plasmid pDG641 (29) using oligos CC009 and CC010.
Plasmid pBS-M5-Kan was then constructed by replacing the ery cassette of pBS-M5-Ery with a kanamycin resistance cassette (kan) amplified from pDG780 (29), using oligos CC011 and CC012. EcoRI–HindIII fragments, containing either the wild-type or mutant rnmV genes fused in frame to a histidine tag, were then excised from the pHMM9-series of plasmids (above) and cloned between EcoRI and HindIII sites located between the rnmV upstream fragment and the kanamycin cassette. The resulting plasmids were called pBS-M5-WT, pBS-M5-353, etc. using the same nomenclature as the pHMM9-series.
By transforming CCB038 with the pBS-M5-mutant series, selecting for kanamycin resistance and screening for erythromycin sensitivity, we were able to integrate both wild-type and mutant rnmV genes onto the chromosome in place of ΔrnmV::ery.
Northern blot analysis of 5S rRNA maturation
Northern blots of 5S rRNA maturation were carried out as described previously (2).
Purification of RNase M5 mutants
Wild-type (WT) and mutant RNase M5 proteins were first applied to His-Trap columns (all columns used in the following purification protocols were from Amersham Biosciences) in 20 mM potassium phosphate buffer, pH 7.4, 250 mM NaCl, 10 mM MgCl2, 5 mM imidazole, 1 mM β-mercapto-ethanol and eluted with a gradient of 5–500 mM imidazole in the same buffer. For the WT and G11A, G61A, I64A, R65A mutants, samples were deemed sufficiently pure after this first column. For the E10A, D56A, D58A and L107A mutants, samples were dialysed against 20 mM Tris, pH 7.2, 50 mM NaCl, 10 mM MgCl2 and 1 mM DTT, applied a MonoS 5/5 column and eluted with a linear gradient of 10 mM–1 M NaCl. For the ΔRRE140–142 mutant, the second purification step was performed on an SP-Sepharose 16/10 column in the same buffer and gradient conditions. For the D56A and ΔRRE140–142 mutants, a third purification step was deemed necessary. The D56A mutant was applied to a Q Fast-Flow column in 20 mM Tris, pH 8, 50 mM NaCl, 10 mM MgCl2, 1 mM DTT and eluted in a 10 mM–1 M NaCl gradient, while the ΔRRE140–142 mutant was applied to a Superdex 75 HR 16/10 column in SBII buffer (10 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 10 mM NH4Cl, 1 mM DTT, 0.1 mM EDTA and 15% glycerol). All proteins were stored in SBII buffer at 1 mg/ml at −80°C.
RNase M5 assays
Unless otherwise stated, RNase M5 activity was assayed in 5 μl reactions in 10 mM Tris pH 7.4, 10 mM MgCl2, 50 mM NH4Cl and 2 mM DTT, containing 250 fmol of labelled 5S precursor, 2.5 μg of yeast competitor RNA (Sigma) and 1 ng of B.subtilis ribosomal protein L18 (2). B.subtilis L18 was purified according to (30), replacing the phosphocellulose columns with MonoS HR (Pharmacia) columns. Reactions in the absence of L18 were performed in the presence of 30% dimethyl sulfoxide (DMSO). Reaction mixes were incubated for 15 min at 37°C, stopped by addition of 5 μl of 95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol and loaded directly on 5% polyacrylamide/7 M urea gels.
Gel retardation assays
An aliquot containing 500 fmol of 32P-labelled 5S rRNA precursor was pre-incubated with 1 pmol B.subtilis L18 in 5 μl binding buffer (30 mM HEPES, pH 7.4, 100 mM NaCl, 0.1 mg/ml BSA, 0.05% Triton X-100 and 0.5 mg/ml yeast tRNA) on ice for 10 min. Aliquots containing 0.5, 1, 2.5, 5 and 10 pmol RNase M5 mutant proteins in 5 μl binding buffer were added to the preformed pre-5S-L18 complex and incubated for a further 15 min on ice. Samples were run on 6% polyacrylamide gels (19:1) in TBE buffer at 20 V/cm for 2.25 h at 4°C. The % RNA bound was quantified from the disappearance of the pre-5S:L18 band.
Purification of Bacillus stearothermophilus RNase M5 and trypsin proteolysis
The B.stearothermophilus rnmV gene was PCR amplified from chromosomal DNA of strain CCM2184 (a gift from Libor Krasny) using oligos HP373 and HP374 and cloned between the EcoRI and HindIII sites of pET21a. The resulting plasmid, pHMM8, was then used as a PCR template to reclone the gene in pET28a using oligos HP461 and HP374, which permitted changing of the Shine-Dalgarno sequence to optimize expression in E.coli. The resulting plasmid, pHMM10, was transformed into E.coli strain BL21 (λDE3) CodonPlus. B.stearothermophilus RNase M5 was purified by applying to an SP Sepharose HP 16/10 column in 50 mM Tris–HCl, pH 7.4, 10 mM MgCl2, 10 mM NH4Cl, 1 mM DTT and elution in a gradient of 10 mM to 1 M NH4Cl. The eluate was applied to a Heparin FF 16/10 column equilibrated in the same buffer and eluted with the same gradient. The protein was concentrated by precipitation with 70% ammonium sulfate and dialysed against SBII buffer for storage at −80°C.
Trypsin digestion was performed at a 1:500 (w/w) ratio of trypsin to RNase M5 (70 μg) in SBII buffer supplemented with 5 mM CaCl2 at 37°C in a 60 μl reaction volume. Five microliter aliquots were withdrawn at the times indicated, and reactions were stopped by the addition of 0.3 μl of 100 mM PMSF and removal to a dry ice/ethanol bath.
RESULTS
The N-terminal half of RNase M5 is related to the Toprim domain
The 5S rRNA processing enzyme, RNase M5, is essentially confined to the low G+C Gram-positive bacteria and the spirochaete Borrelia burgdorferi (2). BLAST searches also revealed a low level of similarity with the archaeal reverse gyrases, bacterial primases and some topoisomerases. These proteins share a fold known as the Toprim domain (6). An alignment of RNase M5 with the Toprim domain of some archaeal reverse gyrases and bacterial topoisomerases and primases is shown in Figure 1. The most highly conserved amino acids cluster around the signature glutamate residue of the N-terminus and the DxD motif of the active site of the Toprim domain.
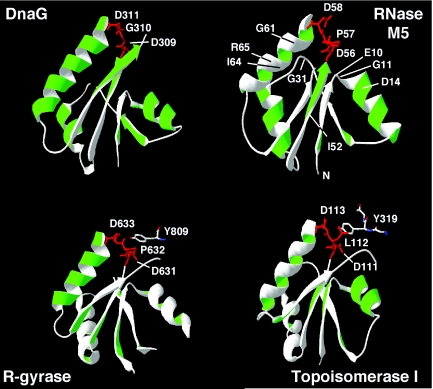
To determine whether the sequence homology would translate to a related structural fold, we performed a PSI-BLAST against the non-redundant database. Six iterations were required before candidates of known structure were identified. The best candidate was the catalytic core of E.coli DNA primase, DnaG [PDB entry 1DD9 (8)], with an E-value of 7 × 10−33 and 23% identity with B.subtilis RNase M5. A model of amino acids Leu6–Pro81 of RNase M5 was built using the structural coordinates of Leu261 to Phe337 of DnaG, as described in Materials and Methods. A comparison of the Toprim domains of E.coli DnaG, A.fulgidus reverse gyrase, E.coli topoisomerase I and the proposed structure of the N-terminal half of RNase M5 is shown in Figure 2.
Figure 2.
Structure of the Toprim domain. Ribbon diagrams of the crystallographic structures of the Toprim domain of E.coli DnaG (PDB code 1DD9; residues 259–341), A.fulgidus reverse gyrase (1GKU; residues 504–524; 544–553; 607–659), E.coli Top1 (1CY0; residues 3–34; 90–141) and a model of the proposed Toprim domain of RNase M5 (residues 6–81). The positions of amino acids mutated in the Toprim domain of RNase M5 are indicated. The DxD motif is shown in red and the positions of the catalytic tyrosine residues of reverse gyrase and topoisomerase are also shown.
Key residues of the Toprim domain are also critical for RNase M5 activity in vivo
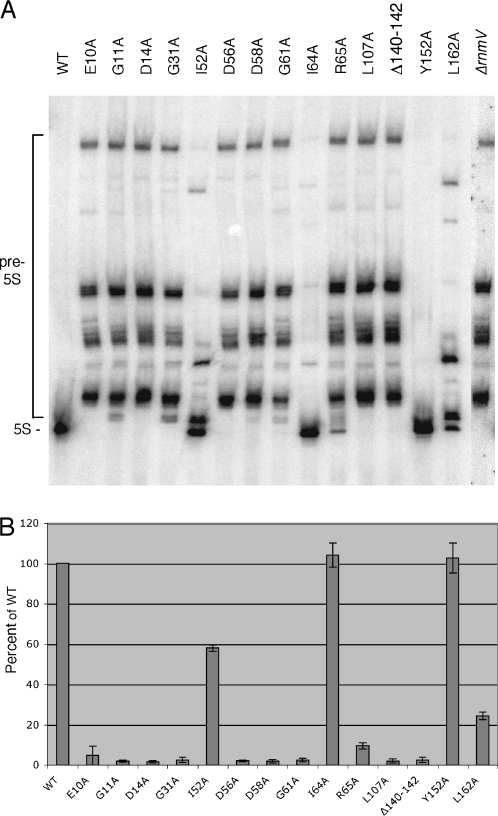
Many of the key residues required for DNA primase and topoisomerase activity are known from previous studies. They are glutamate 9 and the aspartate residues of the DxD motif (D111 and D113) in E.coli TopoI (15,16,31), and glutamate 214 in phage T4 primase (32,33), the equivalent of E265 in DnaG. We hoped to obtain experimental evidence in support of our model by mutation of the equivalent amino acids in RNase M5 and showing they were also important for 5S rRNA maturation. Thirteen of the most highly conserved residues of B.subtilis RNase M5 (10 of them located in the proposed Toprim domain) were mutated and the mutations were introduced onto the chromosome in place of the wild-type rnmV gene. We also deleted residues R140–E142 which are part of an arginine-rich motif in the C-terminal domain that we suspected might be involved in RNA binding. We then determined the effect of these mutations on 5S rRNA maturation in vivo by northern blot (Figure 3A). In the parental strain, only mature 5S rRNA was detected, whereas in a ΔrnmV mutant, all 5S rRNAs were in precursor form, a different precursor for each of the 10 rrn operons of B.subtilis, due to the heterogeneity of their 3′ sequences. The amount of mature 5S rRNA produced varied dramatically with the different rnmV mutations introduced and there were obvious qualitative differences in the precursors that accumulated in some cases. All but three of the mutations introduced in the Toprim domain resulted in a complete inability to produce mature 5S rRNA in vivo. The I52A and I64A mutants had significant or wild-type levels of 5S rRNA maturation, respectively, while the R65A mutant, although not completely inactive, was severely deficient in 5S rRNA production. The relative amounts of mature 5S rRNA produced by each mutant are reported on the histogram shown in Figure 3B. The E10A, G31A, D56A, D58A and G61A mutations, all of which were inactive in vivo, were of particular interest as these residues are fully conserved among the four Toprim domain family members.
Figure 3.
5S rRNA maturation by RNase M5 mutants in vivo. (A) Autoradiogram showing a northern blot of total RNA isolated from the different RNase M5 mutants probed for 5S rRNA. The 5S rRNA precursor (pre-5S) and mature 5S rRNA species (5S) are indicated to the left of the autoradiogram. (B) Histogram showing quantification of the gel shown in (A) and those from two further experiments. Individual precursor species were quantified by PhosphorImager and values represent the percent of the total signal present in the 5S rRNA band, normalized to the wild-type (WT) strain, set at 100%.
Assay of RNase M5 mutants in vitro
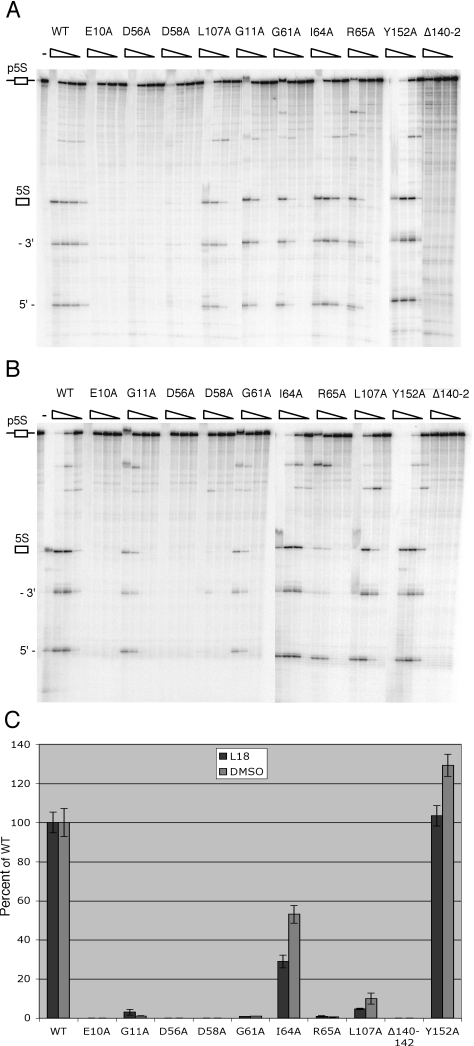
To distinguish between effects on enzyme activity, substrate binding or possible contacts with its co-factor, ribosomal protein L18, histidine-tagged RNase M5 mutants were expressed in E.coli, purified and subjected to 5S rRNA maturation assays in vitro. Four of the mutants (D14A, G31A, I52A and L162A) proved to be unstable, or were not overexpressed in E.coli, and could thus not be purified. The other mutants were purified as described in Materials and Methods and diluted to 1 mg/ml. The specific activity of the different mutants was first compared in 5S rRNA processing assays in which the enzyme was serially diluted 10-fold to 10−3 (Figure 4A). The specific activities relative to wild-type his-tagged RNase M5 are shown in the histogram in Figure 4C. His-tagged RNase M5 has very similar activity to the native protein (data not shown). A very good correlation between the in vivo and in vitro activities was evident for the mutant proteins tested. Mutation of residues E10, D56 and D58, the equivalent of the key active site residues in topoisomerase and DNA primase resulted in a complete (>1000-fold) loss in activity, while the G11A, G61A and R65A mutants were 30- to 100-fold less active than wild-type RNase M5.
Figure 4.
(A) Assay of RNase M5 mutant activities in the presence of ribosomal protein L18. The migration position of the precursor species (p5S), mature 5S RNA (5S), 3′ processed fragment (3′) and 5′ processed fragment (5′) are indicated to the left of the autoradiogram. Mutants were assayed at 200, 20, 2 and 0.2 ng/μl and in the presence of 0.2 ng/μl B.subtilis L18. The direction of the enzyme concentration gradient is indicated by the right-angled triangles at the top of the gel. At the highest enzyme concentration, the precursor band was shifted towards the top of the gel. (B) Assay of RNase M5 mutant activities in the presence of DMSO. Assay conditions were as in (A) except that L18 was replaced by 30% DMSO. (C) Histogram of specific activities of RNase M5 mutants relative to wild-type RNase M5. Wild-type specific activity was 5.17 pmol 5S rRNA generated per μg RNase M5 over the 15 min assay period and was set at 100%. Specific activities were calculated at the lowest dilution of the enzyme to give visible 5S rRNA processing on polyacrylamide gels and are the average of three independent assays.
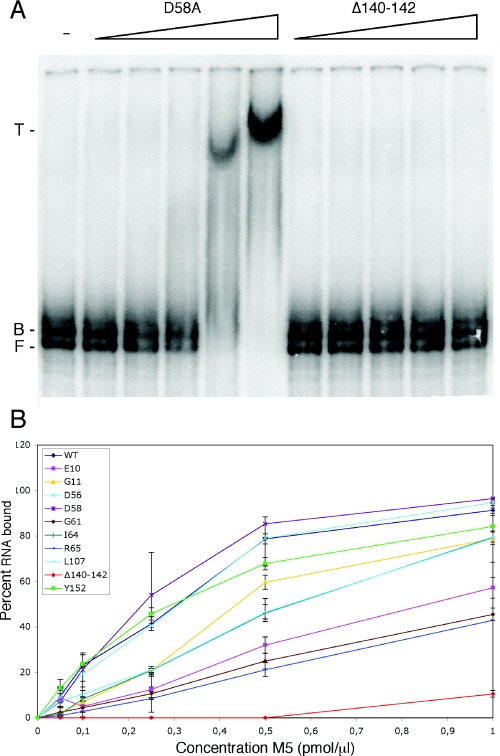
The mutant proteins were then subjected to RNA-binding assays to determine whether they were effected in their ability to bind 5S rRNA precursor. Although RNase M5 can bind 5S rRNA precursor by itself with a Km of ∼2 μM, its affinity is increased by an order of magnitude in the presence of L18 (data not shown). L18 is thought to permit the folding of the 5S rRNA precursor into the correct conformation for cleavage, rather than playing a direct role in the cleavage reaction itself (3). We thus performed gel shift assays of 5S rRNA precursor pre-bound to L18 to determine whether the mutants were affected in their ability to recognize RNA. A gel shift assay showing the most (D58A) and least (Δ140–142) efficient mutants at substrate binding is presented in Figure 5A. Most of the RNase M5 Toprim domain mutants showed at most a 2-fold reduced ability to bind the pre-5S:L18 complex compared with the wild-type protein (Figure 5B), suggesting that a defect in substrate recognition does not account for the dramatic effect of some of these mutations on enzyme activity. In particular, the D56A and D58A mutations in the DxD motif had no effect on RNA-binding capacity, while the E10A mutant retained 50% of the wild-type enzyme's ability to bind RNA.
Figure 5.
Gel retardation assay of RNase M5 mutants. (A) Gel retardation of the 5S rRNA precursor by the D58A and Δ140–142 mutants. The direction of the RNase M5 concentration gradient is indicated by the right-angled triangles. The migration positions of free 5S rRNA precursor (F), the binary complex with ribosomal protein L18 (B) and the ternary complex with L18 and RNase M5 (T) are shown to the left of the autoradiogram. (B) Graph of RNA-binding capacities of RNase M5 mutants and wild-type (WT). Data are the average of three independent experiments.
Although L18 is thought to only play a role of RNA chaperone in the recognition of pre-5S rRNA by RNase M5, the proximity of L18 to the cleavage site (see Discussion) does not preclude the possibility of a direct interaction between RNase M5 and L18. Pace et al. (4) had previously shown that L18 could be functionally replaced in the 5S rRNA processing assay by high concentrations of DMSO and proposed that DMSO facilitated the correct folding of the 5S rRNA precursor. However, effects of DMSO on protein structure have also been demonstrated (34,35). Thus, it is also possible that, in addition to effects on substrate folding, DMSO has an effect on RNase M5 conformation that contributes to its substrate binding or catalytic activity, replacing a similar function of L18. To eliminate the possibility that some of the mutants were affected in an ability to interact directly with L18, we performed 5S rRNA maturation assays where L18 was replaced by 30% DMSO. Under these conditions, wild-type RNase M5 shows equivalent levels of activity to those in the presence of L18 (Figure 4), indicating that, whether L18 affects RNA conformation or RNase M5 conformation (or both), this function can be fully replaced by DMSO. Since the effects of DMSO are likely to be non-specific, i.e. due to global constraints on protein structure, rather acting through specific amino acids characteristic of protein–protein interactions, we reasoned that DMSO should still be able to bypass the requirement for L18 in RNase M5 mutants affected in their ability to interact with it. We thus expected that mutants affected in an interaction with L18 would show near wild-type levels of enzyme activity in the presence of DMSO, whereas mutants affected in catalysis would show similar, decreased levels of RNase M5 activity in the presence of L18 or DMSO. The specific activities of the different mutants in the presence of DMSO corresponded very well with those measured in the presence of L18 (Figure 4), suggesting that the mutations indeed affect enzyme activity rather than a potential interaction with L18.
The Toprim domain of RNase M5 is sensitive to trypsin proteolysis in vitro
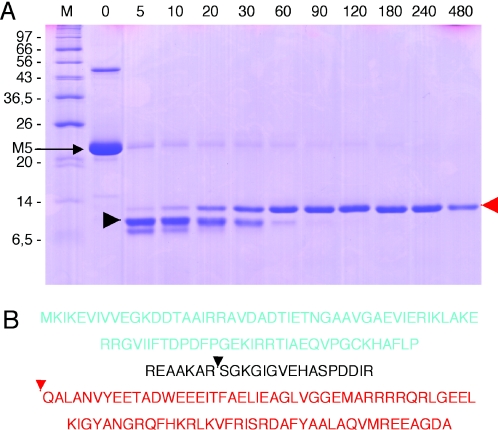
To get further insight into the domain structure of RNase M5, we performed limited trypsin proteolysis of the purified native protein from B.stearothermophilus and ran the products on a 12% polyacrylamide/SDS gel. The B.stearothermophilus protein was used in these experiments with a view to doing subsequent crystallographic studies. After 90 min of incubation, only one proteolysis product was visible. (Figure 6) Mass-spectroscopy and N-terminal sequencing identified this product as the C-terminal 83 amino acids of RNase M5 beginning with the sequence QALAN, suggesting this domain of the protein, but not the N-terminal Toprim domain, is a tightly folded stable domain in vitro. Interestingly, a 100 residue precursor of the C-terminal domain visible at early sampling times migrates faster than the 83 amino acid fragment, suggesting that the 17 additional amino acids cause aberrant migration of the protein due to their charge or structure. No obvious relatives of the C-terminal domain could be identified in the various protein databases.
Figure 6.
Limited trypsin proteolysis of RNase M5. (A) SDS–polyacrylamide gel showing trypsin digestion products. The digestion times are given in minutes above the gel. The migration positions of the molecular weight markers (M) are indicated to the left of the gel. The triangles indicate species analysed by N-terminal sequencing (red) and mass spectroscopy (both red and black). (B) Sequence of B.stearothermophilus RNase M5 indicating the positions of trypsin cleavage. The Toprim domain is in blue lettering; the C-terminal trypsin resistant domain is in red. The positions of the red and black triangles show the N-termini of the corresponding species in (A).
Four mutations were introduced into the C-terminal region of B.subtilis RNase M5 on the basis of their high level of conservation among RNase M5 enzymes. L107A, the third residue after the C-terminal trypsin cleavage site, had a dramatic effect on RNase M5 activity both in vivo and in vitro, while L162A, which was only studied in vivo, had a smaller effect on 5S rRNA maturation (Figures 3 and 4). A deletion of residues Δ140–142 (RRE), which we suspected might play a role in RNA binding due to the similarity of this region with the RNA-binding motif of lambda N protein and HIV Tat, indeed abolished both binding of the L18:pre-5S complex (Figure 5) and enzyme activity (Figures 3 and 4). Thus, these residues are likely to constitute part of the RNA recognition domain. A tyrosine residue plays a key role in the cleavage/religation reaction of E.coli topoisomerase I, performing a nucleophilic attack on the phosphodiester backbone and forming a transient covalent bond with the 5′ end of the downstream cleavage product. Mutation of the only tyrosine residue (Y152A) in B.subtilis RNase M5, however, had no effect on enzyme activity either in vivo or in vitro.
DISCUSSION
In this paper, we show that the N-terminal half of the 5S rRNA maturase can be folded into a Toprim domain structure, characteristic of topoisomerases, primases and reverse gyrases. Mutations of key amino acids for the activity of this family of enzymes also affect RNase M5 activity both in vivo and in vitro. We predict that the mechanism of RNA cleavage by RNase M5 resembles that of DNA cleavage by the topoisomerases, with the catalytic hydroxyl group most likely being provided by water rather than a tyrosine residue. The relationship between the Toprim domains of RNase M5 and topoisomerases supports the notion of a common origin between enzymes acting on RNA and DNA.
The key motifs of the Toprim domain are the DxD motif, thought to coordinate Mg2+ ion, and the glutamate residue (E10 in RNase M5), which has been proposed to play both a role in Mg2+ binding and as a proton donor and in the cleavage reaction by topoisomerases. Mutation of any of these three amino acids completely inactivated B.subtilis RNase M5 both in vivo and in vitro. Mutation of either of the two conserved aspartate residues, D56 and D58, had no effect on RNA binding, while in the case of the E10A mutation, substrate binding was reduced by ∼50%. Interestingly, the equivalent mutation in E.coli topoisomerase I, E9A, also resulted in a 50% reduction in DNA-binding capacity, while the D113A mutation had a more severe effect on substrate binding (75% reduction) than the D58A change in RNase M5 (16). In E.coli topoisomerase I, two Mg++ ions are thought be coordinated by an acidic triad consisting of two aspartate residues (D111 and D113) and glutamate 115 (31). Single mutations of any of these three residues had only a limited effect on topoisomerase relaxation activity, while pairwise mutations or the triple mutation had much more dramatic effects. RNase M5 does not have an equivalent of E115 and thus has one less potential source of Mg++ co-ordination. This may explain the more dramatic effect of the single mutations in the DxD motif in RNase M5 compared with E.coli TopoI. Although the effect of pairwise mutations in the acidic triad of topoisomerase I could be partially rescued by increased Mg2+ concentrations (31), increased Mg2+ concentrations had no effect on the activity of the D56A, D58A or E10A single mutants of RNase M5 (data not shown). Mg2+ is required for a conformational change of E.coli TopoI during the relaxation reaction and is not required for DNA cleavage (36). However, many RNases use Mg2+ as direct participants in the cleavage reaction. For example, in RNase III, the role of Mg2+ is to activate the water nucleophile (37). Although also required for the maturation of 5S rRNA, it is not yet known what role Mg2+ plays in the cleavage by RNase M5—whether it is involved in the cleavage reaction per se, or whether it has a structural function for either the protein or the RNA.
Other mutations in conserved residues of the Toprim domain have effects ranging from a 70% reduction to a 100-fold loss of RNase M5 activity in vitro. Unfortunately, none of the equivalent residues has been studied in other members of the Toprim domain family and thus their function remains unclear. Since most are clustered around the active site it seems likely that they play a role in the correct positioning of the key catalytic residues or of the substrate. Indeed, a complex hydrogen-bond network of residues is involved in the structuring of the catalytic site of E.coli topoisomerase (38). The position of the conserved glycine 31 in the loop between β2 and α2 suggests a role in structure flexibility. Mutation of isoleucines 52 and 64 to alanine had only minor effects on enzyme activity. Our model suggests that these amino acids are involved in hydrophobic interactions within the core of the Toprim domain and this likely explains why they can be replaced by alanine without major deleterious effect. Consistent with this idea, mutation of I52 to arginine and I64 to asparagine had more dramatic effects on RNase M5 activity when compared with the wild-type enzyme in E.coli extracts in vitro (data not shown).
There were qualitative differences in the precursors that accumulated in the different RNase M5 mutants in vivo. This was particularly evident in mutants that were less severely affected in 5S rRNA maturation, e.g. L162A, I64A and I52A. One possible explanation is that the 20 RNase M5 cleavage sites generated from the 10 B.subtilis rrn operons are not all equivalent and that a subset of precursors whose cleavage sites are less well recognized by RNase M5 accumulates in these strains.
DNA cleavage by topoisomerases and reverse gyrases occurs via a nucleophilic attack of the phosphodiester backbone by the hydroxyl group of a highly conserved tyrosine residue. Although a reasonably well-conserved tyrosine residue (Y152) is present in RNase M5, its mutation to alanine had no effect on enzyme activity either in vivo or in vitro. Thus, although the function provided by the Toprim domain is common to the cleavage mechanisms of RNase M5 and topoisomerase I, the nucleophile is different. There appears to be no other residue in the C-terminal half of the protein that is sufficiently highly conserved to provide a hydroxyl function. It is thus likely that nucleophilic attack is performed by a conveniently located water molecule in RNase M5, rather than by the hydroxyl group from an amino acid side chain. Indeed, it has been suggested that residual amounts of activity in the Y319A mutant of E.coli TopoI might be explained by the replacement of nucleophilic function of the tyrosine residue by a solvent water molecule (15).
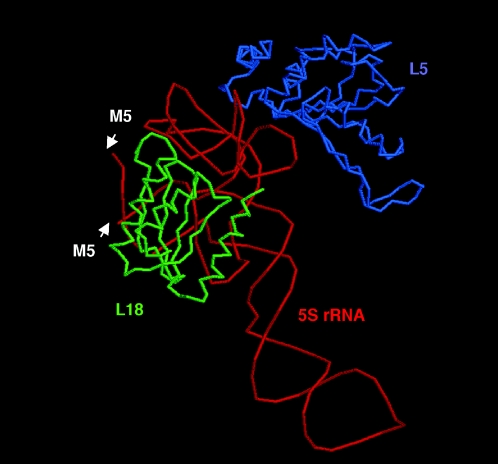
Ribosomal protein L18 is a co-factor in the 5S rRNA maturation reaction in vitro. As shown in Figure 4B, it can be fully replaced by high concentrations of DMSO. It has been suggested that sole function of L18 is to put the 5S rRNA precursor in the correct conformation for cleavage (4). However, the structure of 5S rRNA bound to L18 is known from structural studies of the ribosome (39), and the proximity of ribosomal protein L18 to the mature ends of 5S rRNA (Figure 7) would also permit a direct interaction between RNase M5 and L18. None of the mutants in either the Toprim or C-terminal domain, however, recovered significant levels of activity in the presence of DMSO, suggesting that none of the highly conserved residues at least is involved in such an interaction (or if they are, this does not contribute significantly to enzyme activity). This negative result neither confirms nor excludes the possibility of an interaction between RNase M5 and L18 and it remains possible that other less well-conserved amino acids could be involved in such a role.
Figure 7.
5S rRNA bound to ribosomal proteins L18 and L5 taken from the structure of the D.radiodurans ribosome (PDB code: 1KPJ). The positions of RNase M5 cleavage in the B.subtilis equivalent are indicated by white arrows.
It is difficult to make predictions about the role of residues in the C-terminal half of RNase M5 due to the lack of homology with other proteins. No relationship with the well-known double-stranded RNA processing enzyme, e.g. RNase III, could be seen. The arginine-rich region around residue 140 of RNase M5 is reminiscent of the arginine-rich motif (ARM) of lambda N and HIV Tat proteins involved in RNA binding (40). Deletion of three amino acids from 140–142 (RRE) of B.subtilis RNase M5 completely abolished RNA-binding activity and enzyme activity both in vivo and in vitro, suggesting that this region indeed constitutes an integral part of the substrate binding domain. Confirmation awaits structural studies on the C-terminal domain, as attempts to crystallize the intact protein have so far been unsuccessful.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
The authors thank O. Pellegrini, L. Bénard, M. Springer and P. Stragier for helpful discussion, L. Krasny for B.stearothermophilus chromosomal DNA, J. Dalayer for N-terminal sequencing and J. J. Montagne for mass-spectroscopy analysis. The study was supported by funds from the Centre National de la Recherche Scientifique (UPR 9073), Université de Paris VII-Denis Diderot, PRFMMIP 2003 and ACI Jeunes Chercheurs from the Ministère de l'Education Nationale. Funding to pay the Open Access publication charges for this article was provided by CNRS (UPR9073).
Conflict of interest statement. None declared.
REFERENCES
- 1.Sogin M.L., Pace N.R. In vitro maturation of precursors of 5S ribosomal RNA from Bacillus subtilis. Nature. 1974;252:598–600. doi: 10.1038/252598a0. [DOI] [PubMed] [Google Scholar]
- 2.Condon C., Brechemier-Baey D., Beltchev B., Grunberg-Manago M., Putzer H. Identification of the gene encoding the 5S ribosomal RNA maturase in Bacillus subtilis: mature 5S rRNA is dispensable for ribosome function. RNA. 2001;7:242–253. doi: 10.1017/s1355838201002163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stahl D.A., Pace B., Marsh T., Pace N.R. The ribonucleoprotein substrate for a ribosomal RNA-processing nuclease. J. Biol. Chem. 1984;259:11448–11453. [PubMed] [Google Scholar]
- 4.Pace B., Stahl D.A., Pace N.R. The catalytic element of a ribosomal RNA-processing complex. J. Biol. Chem. 1984;259:11454–11458. [PubMed] [Google Scholar]
- 5.Condon C., Rourera J., Brechemier-Baey D., Putzer H. Ribonuclease M5 has few, if any, mRNA substrates in Bacillus subtilis. J. Bacteriol. 2002;184:2845–2849. doi: 10.1128/JB.184.10.2845-2849.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aravind L., Leipe D.D., Koonin E.V. Toprim—a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 1998;26:4205–4213. doi: 10.1093/nar/26.18.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lima C.D., Wang J.C., Mondragon A. Crystallization of a 67 kDa fragment of Escherichia coli DNA topoisomerase I. J. Mol. Biol. 1993;232:1213–1216. doi: 10.1006/jmbi.1993.1474. [DOI] [PubMed] [Google Scholar]
- 8.Keck J.L., Roche D.D., Lynch A.S., Berger J.M. Structure of the RNA polymerase domain of E.coli primase. Science. 2000;287:2482–2486. doi: 10.1126/science.287.5462.2482. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez A.C., Stock D. Crystal structure of reverse gyrase: insights into the positive supercoiling of DNA. EMBO J. 2002;21:418–426. doi: 10.1093/emboj/21.3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee B.I., Kim K.H., Park S.J., Eom S.H., Song H.K., Suh S.W. Ring-shaped architecture of RecR: implications for its role in homologous recombinational DNA repair. EMBO J. 2004;23:2029–2038. doi: 10.1038/sj.emboj.7600222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiGate R.J., Marians K.J. Escherichia coli topoisomerase III-catalyzed cleavage of RNA. J. Biol. Chem. 1992;267:20532–20535. [PubMed] [Google Scholar]
- 12.Sekiguchi J., Shuman S. Site-specific ribonuclease activity of eukaryotic DNA topoisomerase I. Mol. Cell. 1997;1:89–97. doi: 10.1016/s1097-2765(00)80010-6. [DOI] [PubMed] [Google Scholar]
- 13.Beese L.S., Steitz T.A. Structural basis for the 3′–5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steitz T.A. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 15.Chen S.J., Wang J.C. Identification of active site residues in Escherichia coli DNA topoisomerase I. J. Biol. Chem. 1998;273:6050–6056. doi: 10.1074/jbc.273.11.6050. [DOI] [PubMed] [Google Scholar]
- 16.Zhu C.X., Roche C.J., Papanicolaou N., DiPietrantonio A., Tse-Dinh Y.C. Site-directed mutagenesis of conserved aspartates, glutamates and arginines in the active site region of Escherichia coli DNA topoisomerase I. J. Biol. Chem. 1998;273:8783–8789. doi: 10.1074/jbc.273.15.8783. [DOI] [PubMed] [Google Scholar]
- 17.Kunkel T.A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl Acad. Sci. USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGuffin L.J., Bryson K., Jones D.T. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 20.Gaboriaud C., Bissery V., Benchetrit T., Mornon J.P. Hydrophobic cluster analysis: an efficient new way to compare and analyse amino acid sequences. FEBS Lett. 1987;224:149–155. doi: 10.1016/0014-5793(87)80439-8. [DOI] [PubMed] [Google Scholar]
- 21.Lemesle-Varloot L., Henrissat B., Gaboriaud C., Bissery V., Morgat A., Mornon J.P. Hydrophobic cluster analysis: procedures to derive structural and functional information from 2D representation of protein sequences. Biochimie. 1990;72:555–574. doi: 10.1016/0300-9084(90)90120-6. [DOI] [PubMed] [Google Scholar]
- 22.Douguet D., Labesse G. Easier threading trough web-based comparisons and cross-validation. Bioinformatics. 2001;17:752–753. doi: 10.1093/bioinformatics/17.8.752. [DOI] [PubMed] [Google Scholar]
- 23.Kelley L.A., MacCallum R.M., Sternberg M.J.E. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- 24.Jones D.T. GenTHREADER: an efficient and reliable protein fold recognition method for genomic sequences. J. Mol. Biol. 1999;287:797–815. doi: 10.1006/jmbi.1999.2583. [DOI] [PubMed] [Google Scholar]
- 25.Shi J., Blundell T.L., Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 2001;310:243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- 26.Labesse G., Mornon J.P. Incremental threading optimization (TITO) to help alignment and modelling of remote homologues. Bioinformatics. 1998;14:206–211. doi: 10.1093/bioinformatics/14.2.206. [DOI] [PubMed] [Google Scholar]
- 27.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 28.Sippl M.J. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993;17:355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- 29.Guerout-Fleury A.M., Shazand K., Frandsen N., Stragier P. Antibiotic-resistance cassettes for Bacillus subtilis. Gene. 1995;167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- 30.Pace N.R., Pace B. Ribosomal RNA terminal maturase: ribonuclease M5 from Bacillus subtilis. Methods Enzymol. 1990;181:366–374. doi: 10.1016/0076-6879(90)81136-i. [DOI] [PubMed] [Google Scholar]
- 31.Zhu C.X., Tse-Dinh Y.C. The acidic triad conserved in type IA DNA topoisomerases is required for binding of Mg(II) and subsequent conformational change. J. Biol. Chem. 2000;275:5318–5322. doi: 10.1074/jbc.275.8.5318. [DOI] [PubMed] [Google Scholar]
- 32.Strack B., Lessl M., Calendar R., Lanka E. A common sequence motif, -E-G-Y-A-T-A-, identified within the primase domains of plasmid-encoded I- and P-type DNA primases and the alpha protein of the Escherichia coli satellite phage P4. J. Biol. Chem. 1992;267:13062–13072. [PubMed] [Google Scholar]
- 33.Ziegelin G., Linderoth N.A., Calendar R., Lanka E. Domain structure of phage P4 alpha protein deduced by mutational analysis. J. Bacteriol. 1995;177:4333–4341. doi: 10.1128/jb.177.15.4333-4341.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pande C., Callender R., Henderson R., Pande A. Purple membrane: color, crystallinity, and the effect of dimethyl sulfoxide. Biochemistry. 1989;28:5971–5978. doi: 10.1021/bi00440a038. [DOI] [PubMed] [Google Scholar]
- 35.Bhattacharjya S., Balaram P. Effects of organic solvents on protein structures: observation of a structured helical core in hen egg-white lysozyme in aqueous dimethylsulfoxide. Proteins. 1997;29:492–507. doi: 10.1002/(sici)1097-0134(199712)29:4<492::aid-prot9>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 36.Zhu C.X., Roche C.J., Tse-Dinh Y.C. Effect of Mg(II) binding on the structure and activity of Escherichia coli DNA topoisomerase I. J. Biol. Chem. 1997;272:16206–16210. doi: 10.1074/jbc.272.26.16206. [DOI] [PubMed] [Google Scholar]
- 37.Sun W., Pertzev A., Nicholson A.W. Catalytic mechanism of Escherichia coli ribonuclease III: kinetic and inhibitor evidence for the involvement of two magnesium ions in RNA phosphodiester hydrolysis. Nucleic Acids Res. 2005;33:807–815. doi: 10.1093/nar/gki197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lima C.D., Wang J.C., Mondragon A. Three-dimensional structure of the 67K N-terminal fragment of E.coli DNA topoisomerase I. Nature. 1994;367:138–146. doi: 10.1038/367138a0. [DOI] [PubMed] [Google Scholar]
- 39.Harms J., Schluenzen F., Zarivach R., Bashan A., Gat S., Agmon I., Bartels H., Franceschi F., Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 40.Lazinski D., Grzadzielska E., Das A. Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell. 1989;59:207–218. doi: 10.1016/0092-8674(89)90882-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.