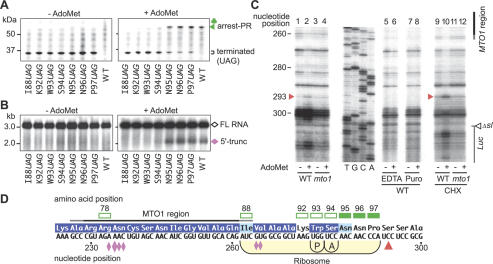
Figure 4.
Determination of the position of the translation elongation arrest. (A,B) Effects of stop codon substitutions. GST:Ex1:Luc RNAs carrying the stop codon substitution constructs were translated for 30 min (A) or 90 min (B) in the presence (right) or absence (left) of AdoMet. Aliquots were subjected to immunoblot analysis with anti-GST antibody (A) or Northern blot analysis with LUC 3′ probe (B). The full-length polypeptide (106 kDa) is not seen in A. Brackets in A mark the complete translation products terminated at the introduced stop codon. (C) Toeprint analysis. GST:Ex1Δsl(WT):Luc RNA (lanes 1,2,5-10) or GST:Ex1Δsl(mto1-1):Luc RNA (lanes 3,4,11,12) was translated for 30 min in the presence (+) or absence (-) of AdoMet. In lanes 5 and 6, EDTA was added to a final concentration of 5 mM at 30 min, and the samples were incubated for another 5 min. MgCl2 was added to 10 mM prior to the primer extension reaction. In lanes 7 and 8, puromycin (Puro) was added to 1 mM at 30 min, and the reaction mixtures were incubated for an additional 10 min. In lanes 9-12, 0.5 mg/mL CHX was added at 30 min. The red arrowheads mark the most 5′-proximal AdoMet-dependent toeprint signals at the 293rd nucleotide position. The sequence ladder (shown in the sense strand sequence) was synthesized using the same primer as used for toeprinting. The MTO1 region and the Δsl:Luc junction are indicated. (D) Schematic representation of the translation elongation arrest position. The red arrowhead indicates the position of the most 5′-proximal AdoMet-dependent toeprint signal. The positions of the P and A sites of the stalled ribosome deduced from the toeprint signal are shown. Open squares indicate codons whose substitution to a stop codon abolished production of the AdoMet-dependent peptidyl-tRNA and 5′-truncated RNA, while solid squares indicate codons without any effect. Amino acid residues that are conserved among plant CGSs are reversed in purple and those with conservative changes are shaded in light blue (Ominato et al. 2002). The pink diamonds indicate the positions of the 5′ ends of the 5′-truncated RNA species with the large one pointing to the most prominent one (Chiba et al. 2003). Representative results of triplicate to quadruplicate experiments are shown.