Abstract
Background
The alpha (α) proteobacteria, a very large and diverse group, are presently characterized solely on the basis of 16S rRNA trees, with no known molecular characteristic that is unique to this group. The genomes of three α-proteobacteria, Rickettsia prowazekii (RP), Caulobacter crescentus (CC) and Bartonella quintana (BQ), were analyzed in order to search for proteins that are unique to this group.
Results
Blast analyses of protein sequences from the above genomes have led to the identification of 61 proteins which are distinctive characteristics of α-proteobacteria and are generally not found in any other bacteria. These α-proteobacterial signature proteins are generally of hypothetical functions and they can be classified as follows: (i) Six proteins (CC2102, CC3292, CC3319, CC1887, CC1725 and CC1365) which are uniquely present in most sequenced α-proteobacterial genomes; (ii) Ten proteins (CC1211, CC1886, CC2245, CC3470, CC0520, CC0365, CC0366, CC1977, CC3010 and CC0100) which are present in all α-proteobacteria except the Rickettsiales; (iii) Five proteins (CC2345, CC3115, CC3401, CC3467 and CC1021) not found in the intracellular bacteria belonging to the order Rickettsiales and the Bartonellaceae family; (iv) Four proteins (CC1652, CC2247, CC3295 and CC1035) that are absent from various Rickettsiales as well as Rhodobacterales; (v) Three proteins (RP104, RP105 and RP106) that are unique to the order Rickettsiales and four proteins (RP766, RP192, RP030 and RP187) which are specific for the Rickettsiaceae family; (vi) Six proteins (BQ00140, BQ00720, BQ03880, BQ12030, BQ07670 and BQ11900) which are specific to the order Rhizobiales; (vii) Four proteins (BQ01660, BQ02450, BQ03770 and BQ13470) which are specific for the order Rhizobiales excluding the family Bradyrhizobiaceae; (viii) Nine proteins (BQ12190, BQ11460, BQ11450, BQ11430, BQ11380, BQ11160, BQ11120, BQ11100 and BQ11030 which are distinctive of the Bartonellaceae family;(ix) Six proteins (CC0189, CC0569, CC0331, CC0349, CC2323 and CC2637) which show sporadic distribution in α-proteobacteria, (x) Four proteins (CC2585, CC0226, CC2790 and RP382) in which lateral gene transfers are indicated to have occurred between α-proteobacteria and a limited number of other bacteria.
Conclusion
The identified proteins provide novel means for defining and identifying the α-proteobacteria and many of its subgroups in clear molecular terms and in understanding the evolution of this group of species. These signature proteins, together with the large number of α-proteobacteria specific indels that have recently been identified http://www.bacterialphylogeny.com, provide evidence that all species from this diverse group share many unifying and distinctive characteristics. Functional studies on these proteins should prove very helpful in the identification of such characteristics.
Background
The α-proteobacteria comprise a large and extremely diverse group of Gram-negative bacteria which form a part of the largest known phyla within prokaryotes, namely the proteobacteria [1]. The vast diversity of the α-subdivision is clearly evident through the lifestyle differences among its members making them important in agricultural, medical and industrial fields. Such examples include the animal and human intracellular pathogens (Rickettsia, Bartonella, and Brucella) [1-3], the plant pathogens and symbiotic soil bacteria (Agrobacterium, Sinorhizobium, Mesorhizobium, and Bradyrhizobium) [1,4-6], the Drosophila endosymbiont (Wolbachia) [1] and a number of other free-living bacteria occupying a wide variety of ecological niches [1]. Furthermore, this group exhibits a wide spectrum of characteristics in terms of morphology (spiral, rod, stalked), metabolism (phototrophs, heterotrophs, and chemolithotrophs), physiology and cell division mechanisms [1,7,8]. In addition to their great diversity in these regards, this group of species is also of central importance due to compelling evidence indicating that a large proportion of the genes in eukaryotic cells, especially those related to mitochondria, have an α-proteobacterial ancestry [9-16].
In the current view, the α-subdivision are thought to form a more recently branching monophyletic taxon emerging after the epsilon and delta but before the beta and gamma subdivisions or Classes of proteobacteria [1,13,17]. Although this group is distinguished from other major bacterial groups based on 16S rRNA and other gene phylogenies [7,13,17-19], no set of criteria exists to clearly define and circumscribe the α-proteobacteria in clear and unambiguous molecular terms [1]. Thus, the following question remains: what defining molecular characteristics distinguish an α-proteobacterium and its subgroups from all other bacteria? The task of identifying such markers is aided by the availability of 18 completely sequenced α-proteobacterial genomes along with 10 partially sequenced genomes [11,20-33], belonging to the following orders: Rhizobiales, Rickettsiales, Caulobacterales, Rhodobacterales, Sphingomonadales and Rhodospirillales [34]. The comparative analyses of genomes provides a valuable resource and a very powerful means for identifying characteristics that are unique to a particular group of species [6,16,27,28,32,35,36]. We have used these data to identify a large number of conserved inserts and deletions (indels) in protein sequences that are distinctive characteristics of different groups of bacteria and provide molecular means for their identification and characterization [13,37-40]. Recently, we have also identified many conserved indels in protein sequences that are useful for defining the α-proteobacteria group, and its various subgroups, in molecular terms [17]. The distribution pattern of these signatures in different α-proteobacteria has been used to deduce a working model to describe the interrelationships as well as the branching order among the α-proteobacteria species [17].
In the present study, a new type of taxonomic marker is described which provides an additional means to define the α-proteobacteria group as well as the relationship within this group. These new markers consist of whole proteins that are specific to certain groups or subgroups of bacteria and are not found in any other phyla [35]. In this work we have identified a large number of proteins which are specific to either the α-proteobacteria group as a whole or its various subgroups. These signature proteins were identified in BLASTP searches [41] of individual proteins from the genomes of three α-proteobacterial species (viz. Rickettsia prowazekii, Caulobacter crescentus and Bartonella quintana) [11,24,32], which show important differences in lifestyles and physiology. Results of this study presented here will prove useful in developing a clearer picture of α-proteobacterial phylogeny as well as aid in the identification of bacterial strains belonging to this group and its subgroups. Functional studies on these α-proteobacteria specific proteins should prove instrumental in the discovery of novel physiological characteristics that are uniquely shared by members of this large and diverse group of bacteria.
Results
These studies were undertaken with the aim of identifying proteins that are uniquely found in α-proteobacteria and which could provide novel molecular means for defining and identifying bacteria belonging to this group and its subgroups. To identify proteins which are specific to α-proteobacteria or its subgroups, BLAST searches were carried out individually on every single annotated protein present in the genomes of three different α-proteobacteria, C. crescentus, R. prowazekii and B. quintana. These genomes were chosen because of their different sizes (R. prowazekii, 1.11 Mb with 835 open reading frames (ORFs); B. qunitana, 1.58 Mb, 1142 ORFs; C. crescentus, 4.02 Mb with 3737 ORFs) and because these species display important differences in life-style and other characteristics [11,24,32]. Results of the BLAST searches were inspected in order to identify proteins which are only found in α-proteobacteria, as well as proteins where the only acceptable BLAST scores as indicated by their expected values (E values) were from α-proteobacteria [41]. These studies have resulted in the identification of 61 signature proteins, which appear distinctive of α-proteobacteria and are generally not found in any other Bacteria. For all of these proteins, the lengths of the query proteins as well as the E values obtained from BLAST searches for different hits are shown (Tables 1, 2, 3, 4, 5, 6, 7, 8, 9). The former values are important in determining the significance of the observed BLAST scores (See Methods section). Additionally, for all of the α-proteobacteria specific proteins, the length of the hit protein over the query sequence is shown in brackets to show that the homologues in different species are of similar length. Most of the α-proteobacterial signature proteins that we have identified are of hypothetical function as annotated in the NCBI database http://www.ncbi.nlm.nih.gov/genomes/MICROBES/Complete.html. For the sake of presentation and discussion, we have arbitrarily divided these proteins into ten groups based on their distribution patterns among α-proteobacteria.
Table 1.
Signature Proteins Specific for the Alpha-proteobacteria.a
| Protein | CC2102 [16126341] | CC3292 [16127522] | CC3319b [16127549] | CC1887c [16126130] | CC1725 [16125969] | CC1365 [16125614] |
| Length | 162 | 224 | 89 | 105 | 100 | 161 |
| Mag. mag. | 2e-19 (1.04) | 9e-49 (1.29) | 2e-15 (0.99) | 3e-20 (1.10) | 1e-08 (0.88) | 2e-21 (0.87) |
| Rhod. rubr. | 3e-17 (1.09) | 1e-42 (0.92) | 1e-12 (1.04) | 4e-21 (1.12) | 1e-08 (1.09) | 3e-17 (0.96) |
| Nov. aro. | 1e-19 (1.02) | 3e-45 (1.04) | 4e-07 (0.88) | 2e-25 (1.30) | - | 1e-11 (0.98) |
| Z. mobilis * | 4e-24 (1.01) | 6e-40 (1.00) | - | - | - | 5e-11 (1.02) |
| C. cres.* | 1e-64 (1.00) | e-106 (1.00) | 1e-34 (1.00) | 3e-61 (1.00) | 4e-49 (1.00) | 7e-79 (1.00) |
| Sil. pom.* | 3e-16 (0.93) | 2e-44 (1.15) | 8e-11 (0.97) | 3e-22 (1.21) | 8e-06 (0.99) | 4e-12 (0.96) |
| Sil. sp. | 1e-13 (0.98) | 1e-43 (1.14) | 8e-09 (0.99) | 5e-24 (1.22) | 1e-05 (0.88) | 9e-12 (0.89) |
| Rh. spha. | 1e-16 (0.94) | 3e-43 (1.11) | 4e-08 (0.94) | 8e-24 (1.18) | 2e-08 (1.00) | 1e-11 (0.91) |
| Bra. jap.* | 5e-21 (1.06) | 8e-47 (1.11) | 2e-12 (1.01) | 2e-29 (1.29) | 2e-09 (1.05) | 2e-11 (1.15) |
| Rho. pal.* | 8e-22 (1.10) | 4e-45 (1.06) | 7e-13 (1.45) | 2e-25 (1.30) | 2e-08 (1.05) | 2e-12 (1.00) |
| Agr. tum.* | 4e-21 (1.04) | 7e-49 (1.02) | 4e-10 (0.96) | 3e-26 (1.26) | 5e-06 (1.14) | 7e-13 (1.10) |
| Sino. meli. * | 4e-23 (1.06) | 2e-49 (1.04) | 6e-12 (1.01) | 2e-22 (1.25) | 5e-05 (0.93) | 1e-14 (1.04) |
| Bru. mel.* | 5e-19 (1.17) | 4e-48 (1.02) | 1e-12 (1.03) | 1e-23 (1.30) | 1e-07 (1.09) | 4e-15 (1.04) |
| Bru. suis* | 5e-19 (1.17) | 4e-48 (1.02) | 2e-12 (0.97) | 1e-23 (1.30) | 2e-08 (1.09) | 4e-15 (1.07) |
| Meso. loti* | 5e-18 (1.10) | 9e-49 (1.00) | 9e-18 (0.97) | 9e-23 (1.26) | 4e-09 (1.20) | 3e-14 (1.40) |
| Meso. sp. | 2e-17 (1.09) | 2e-47 (1.03) | 5e-16 (0.98) | 6e-27 (1.26) | 8e-06 (1.05) | 7e-15 (1.33) |
| B. henselae* | 8e-14 (1.12) | 5e-41 (1.04) | 4e-12 (0.96) | 2e-23 (1.26) | 5e-08 (1.09) | 4e-10 (0.99) |
| B. Quintana* | 5e-14 (1.12) | 8e-42 (1.02) | 4e-12 (0.96) | 5e-21 (1.26) | 1e-08 (1.09) | 3e-11 (0.99) |
| R. conorii* | 5e-10 (0.97) | 3e-41 (0.83) | 1e-07 (0.88) | 1e-05 (0.94) | 5e-11 (1.01) | 7e-07 (0.98) |
| R. prowazekii* | 4e-08 (0.98) | 4e-40 (0.83) | 3e-07 (0.88) | 5e-07 (0.95) | 2e-12 (1.07) | 8e-06 (0.95) |
| R. typhi* | 1e-08 (0.98) | 2e-40 (0.83) | 3e-07 (0.88) | 4e-07 (0.95) | 5e-11 (1.07) | 3e-06 (0.98) |
| R. akari | 6e-09 (0.97) | 9e-41 (0.90) | 1e-07 (0.88) | 2e-05 (0.94) | 7e-12 (1.07) | 3e-07 (0.98) |
| R. rickettsii | 1e-10 (0.97) | 9e-41 (0.83) | 1e-07 (0.88) | 1e-05 (0.94) | 5e-11 (1.01) | 7e-07 (0.98) |
| R. sibirica | 2e-10 (0.97) | 5e-41 (0.83) | 1e-07 (0.88) | 1e-05 (0.94) | 5e-11 (1.01) | 3e-07 (0.98) |
| Wolbachia* | 5e-11 (0.90) | 3e-20 (0.89) | 2e-07 (0.99) | 1e-08 (0.99) | 1e-05 (1.04) | 3e-04 (0.70) |
| Ana. mar.* | 2e-09 (0.98) | 2e-40 (0.92) | - | - | 9e-07 (1.19) | - |
| Ehr. canis | 2e-07 (1.01) | 6e-39 (0.95) | 2e-06 (1.04) | 5e-10 (0.94) | - | 0.023 (0.90) |
| Ehr. rum.* | 1e-08 (0.94) | 2e-39 (0.99) | 2e-05 (1.07) | 8e-11 (0.89) | - | 4e-05 (0.84) |
| Non-Alpha | - | Strep. glau. 1.6 (1.89) | Fuso. nucl. 2.9 (3.87) | Burk. fung. (3.69) | Azo. sp. 0.44 (0.91) | Myc. pneum. 0.98 (2.63) |
a Alpha-specific proteins were identified by BLAST searches on individual protein sequences on three α-proteobacterial genomes as described in the Methods section. The expected (E) values for various alpha-proteobacteria species as well as the first non-alpha species in the BLAST results are shown here. The values in brackets after the E values represent the ratios of the length of the hit protein divided by the query protein and a value close to 1.0 indicates that the homologues are of similar lengths. The CC numbers indicate the protein identification number in the C. crescentus genome. GenBank accession numbers for the query sequence are shown in square brackets. An asterisk (*) identifies bacterial genomes which are completely sequenced, whereas other sequences are from partially or incompletely sequenced genomes. Proteins not found in a given species are indicated with a dash (-). Abbreviations: Agr. tum., Agrobacterium tumefaciens; Ana. mar., Anaplasma marginale; Azo. sp., Azoarcus sp. EbN1; B. henselae, Bartonella henselae; B. quintana, B. quintana; Bra. jap., Bradyrhizobium japonicum; Bru. mel., Brucella melitensis; Burk. fung., Burkholderia fungorum; C. cres., Caulobacter crescentus; Ehr. canis, Ehrlichia canis; Ehr. rum., Ehr ruminantium; Fuso. nucl., Fusobacterium nucleatum; Mag. mag., Magnetospirillum magnetotacticum; Meso., Mesorhizobium; Myc. pneum., Mycoplasma pneumoniae; Nov. aro., Novosphingobium aromaticivorans; Rhod. rubr., Rhodospirillum rubrum; Rh. spha., Rhodobacter sphaeroides; Rho. pal., Rhodopseudomonas palustris; Sil. pom., Silicibacter pomeroyi; Sil. sp., Silicibacter sp. TM1040; Sino. meli., Sinorhizobium meliloti; Strep. glau., Streptomyces glaucescens; Wolbachia, Wolbachia endosymbiont of Drosophila melanogaster.
b Magnetococcus sp. MC-1 (unclassfied) found in BLAST search with E value of 3e-09 [48832993].
c Protein also found in Eukaryotes. The E values for a few representative eukaryotic species are as follows: Homo sapiens; 8e-10 [30583279], Chlamydomonas reinhardtii; 3e-09 [34334022], Caenorhabditis elegans; 5e-06 [7332202].
Table 2.
Signature Proteins specific for Alpha-proteobacteria, except Rickettsiales.a
| Protein | CC1211 [16125461] | CC1886 [16126129] | CC2245 [16126484] | CC3470 [16127700] | CC0520b [16124775] |
| Length | 167 | 223 | 190 | 253 | 284 |
| Mag. mag. | 3e-19 (1.09) | - | 3e-16 (0.96) | 5e-23 (0.97) | 2e-38 (0.89) |
| Rhod. rubr. | 3e-17 (0.73) | 1e-07 (0.73) | 3e-22 (0.64) | 5e-04 (0.82) | 2e-36 (0.93) |
| Nov. aro. | 9e-13 (1.41) | 3e-05 (0.91) | 7e-13 (1.24) | 6e-20 (0.84) | - |
| Z. mobilis | 8e-12 (1.38) | - | 5e-12 (1.21) | 1e-15 (0.82) | - |
| C. cres. | 5e-93 (1.00) | 3e-58 (1.00) | 1e-75 (1.00) | e-115 (1.00) | e-146 (1.00) |
| Sil. pom. | 9e-21 (1.19) | 2e-04 (0.53 | 7e-16 (1.04) | 2e-10 (0.78) | 5e-27 (0.88) |
| Sil. sp. | 8e-20 (1.17) | 5e-07 (0.65) | 1e-15 (1.03) | 3e-07 (0.78) | 1e-27 (0.87) |
| Rh. spha. | 1e-19 (1.24) | 2e-06 (0.55) | 2e-16 (1.09) | 8e-07 (0.78) | 2e-30 (0.92) |
| Bra. jap. | 4e-20 (1.01) | 1e-10 (1.61) | 1e-25 (0.89) | 5e-17 (0.94) | 2e-44 (0.87) |
| Rho. pal. | 1e-18 (1.28) | 5e-10 (1.48) | 3e-25 (1.12) | 2e-17 (0.86) | 5e-45 (0.91) |
| Agr. tum. | - | 2e-10 (0.65) | - | 1e-15 (0.85) | 8e-46 (1.01) |
| Sino. meli. | 1e-19 (1.04) | 3e-12 (0.67) | 4e-25 (0.91) | 7e-16 (0.87) | 1e-45 (0.89) |
| Bru. mel. | 4e-22 (1.11) | 2e-10 (0.68) | 1e-26 (0.97) | 1e-18 (0.82) | 2e-41 (1.08) |
| Bru. suis | 4e-22 (1.08) | 2e-10 (0.70) | 5e-26 (0.95) | 8e-18 (0.83) | 1e-41 (0.91) |
| Meso. loti | 4e-24 (1.02) | 5e-09 (0.89) | 1e-27 (0.90) | 8e-21 (0.83) | 1e-44 (0.92) |
| Meso. sp. | 1e-21 (1.07) | 2e-10 (0.55) | 5e-25 (0.94) | 1e-18 (0.83) | 1e-42 (0.91) |
| B. henselae | 6e-20 (1.06) | 9e-14 (0.63) | 9e-22 (0.93) | 5e-15 (0.84) | 1e-28 (0.86) |
| B. quintana | 2e-19 (1.06) | 3e-12 (0.63) | 3e-22 (0.93) | 2e-11 (0.84) | 2e-27 (0.89) |
| Non Alpha | Kin. radio. 0.097 (3.89) | Vib. para. 7.7 (0.82) | Pse. aeruginosa 4.2 (1.88) | Vib. cholerae 0.035 (1.96) | Polaromonas sp. 0.003 (0.95) |
| Protein | CC0365 [16124620] | CC0366c [16124621] | CC1977d [16126220] | CC3010e [16127240] | CC0100 [16124355] |
| Length | 169 | 177 | 241 | 216 | 576 |
| Mag. mag. | 2e-06 (1.05) | 2e-06 (0.93) | 1e-32 (0.98) | 4e-24 (0.90) | 7e-34 (0.92) |
| Rhod. rubr. | 2e-08 (1.08) | 4e-06 (0.91) | 2e-36 (1.07) | 8e-18 (0.94) | - |
| Nov. aro. | 3e-07 (1.09) | 3e-05 (0.93) | 5e-29 (0.95) | 3e-13 (0.91) | 5e-26 (1.09) |
| Z. mobilis | - | - | 1e-35 (0.97) | 6e-12 (0.96) | 2e-35 (1.02) |
| C. cres. | 1e-44 (1.00) | 1e-44 (1.00) | e-111 (1.00) | 6e-93 (1.00) | 0.0 (1.00) |
| Sil. pom. | 4e-09 (1.12) | 1e-07 (1.02) | 2e-40 (0.99) | 1e-19 (0.95) | 2e-34 (0.99) |
| Sil. sp. | 4e-08 (1.09) | 1e-11 (1.02) | 6e-38 (0.97) | 2e-18 (0.96) | 1e-33 (1.00) |
| Rh. spha. | 1e-06 (1.09) | 1e-08 (0.90) | 6e-36 (0.98) | 2e-20 (0.98) | 7e-30 (0.98) |
| Bra. jap. | 8e-07 (0.95) | 2e-12 (1.06) | 1e-28 (1.10) | 8e-20 (0.94) | 3e-35 (1.02) |
| Rho. pal. | 9e-06 (0.96) | 9e-10 (1.05) | 2e-33 (1.08) | 2e-19 (0.99) | 5e-35 (1.09) |
| Agr. tum. | 1e-06 (0.95) | 7e-10 (1.21) | 2e-35 (1.10) | 3e-20 (0.98) | 8e-35 (0.98) |
| Sino. meli. | 2e-08 (0.95) | 4e-10 (1.15) | 3e-38 (1.08) | 2e-20 (0.84) | 2e-31 (0.94) |
| Bru. mel. | 0.002 (0.84) | 1e-10 (1.02) | 1e-32 (1.08) | 4e-20 (0.97) | 4e-36 (0.99) |
| Bru. suis | 2e-07 (0.94) | 1e-10 (1.18) | 8e-33 (1.08) | 4e-20 (0.97) | 5e-36 (0.94) |
| Meso. loti | 1e-08 (0.96) | 8e-08 (1.09) | - | 5e-22 (0.95) | 2e-34 (0.92) |
| Meso. sp. | 7e-11 (0.94) | 5e-11 (1.09) | 1e-27 (1.09) | 6e-23 (0.95) | 1e-34 (0.99) |
| B. henselae | 1e-06 (0.97) | 3e-08 (1.06) | 1e-27 (1.08) | 2e-20 (0.95) | 2e-27 (0.99) |
| B. quintana | 5e-07 (0.97) | 1e-07 (1.06) | 2e-29 (1.08) | 2e-20 (0.94) | 4e-30 (0.99) |
| Non-Alpha | Myc. galli. 1.1 (1.17) | Rhodo. baltica 0.005 (1.47) | M. thermo. 0.28 (1.03) | Syn. elongatus 0.002 (1.33) | Coryn. efficiens 0.11 (0.52) |
a Abbreviations and other details regarding BLAST results can be found in Table 1. E values of 0.0 indicate an extremely high degree of similarity between protein sequences. Additional abbreviations: Coryn., Corynebacterium; Kin. radio., Kineococcus radiotolerans; M. thermo., Moorella thermoacetica; Myc. galli., Mycoplasma gallisepticum; Pse., Pseudomonas; Rhodo., Rhodopirellula; Syn., Synechococcus; Vib. para., Vibrio parahaemolyticus.
b,c Magnetococcus sp. MC-1 (unclassified) contains homologues of both proteins with E values of 1e-18 [48832519] and 8e-06 [48833234] respectively.
d Protein also found in Eukaryotes with examples from representative species as follows: Homo sapiens; 1e-22 [21735485], Oryza sativa; 2e-18 [50939575] and Cryptococcus neoformans; 3e-17 [57225838].
e One BLAST hit is Pseudomonas sp. with E value of 8e-20 [94976].
Table 3.
Alpha-proteobacteria specific proteins which are absent in the Rickettsiales as well as (A) the Bartonellaceae family, or (B)the Rhodobacterales a
| Protein | CC2345 [16126584] | CC3115 [16127345] | CC3401 [16127631] | CC3467 [16127697] | CC1021 [16125273] |
| Length | 159 | 136 | 120 | 152 | 130 |
| Mag. mag. | 2e-38 (0.99) | 2e-29 (0.90) | 3e-14 (1.03) | 7e-16 0.86 | 3e-14 (1.16) |
| Rhod. rubr. | 1e-39 (1.01) | - | - | - | - |
| Nov. aro. | 4e-35 (1.03) | 8e-32 (1.08) | 1e-09 (1.23) | 1e-19 (1.10) | 5e-06 (1.13) |
| C. cres. | 2e-86 (1.00) | 9e-79 (1.00) | 8e-56 (1.00) | 3e-82 (1.00) | 1e-57 (1.00) |
| Sil. pom. | 2e-38 (1.01) | 1e-09 (0.96) | 3e-09 (1.08) | 5e-23 (0.99) | 3e-14 (1.00) |
| Sil. sp. | 3e-40 (1.00) | 5e-10 (1.05) | 5e-10 (1.19) | 5e-24 (1.34) | 8e-16 (1.10) |
| Rh. spha. | 6e-40 (0.99) | 1e-07 (1.00) | 9e-07 (1.13) | 1e-25 (1.02) | 4e-10 (1.05) |
| Bra. jap. | 2e-44 (1.04) | 4e-27 (0.98) | 6e-11 (1.22) | 5e-28 (1.07) | 6e-17 (1.12) |
| Rho. pal. | 2e-45 (1.03) | 8e-24 (0.96) | 5e-13 (1.09) | 5e-27 (1.05) | 9e-15 (1.09) |
| Agr. tum. | 7e-44 (0.99) | 6e-30 (0.94) | 2e-15 (1.07) | 3e-28 (1.06) | 1e-10 (0.98) |
| Sino. meli. | 3e-44 (0.99) | 1e-25 (0.83) | 4e-14 (1.06) | 7e-30 (1.14) | 7e-13 (1.16) |
| Bru. mel. | 2e-43 (1.01) | 1e-28 (1.12) | 3e-15 (1.27) | 9e-29 (1.08) | 1e-10 (1.17) |
| Bru. suis | 2e-43 (1.01) | 4e-29 (0.93) | 2e-15 (1.06) | 9e-29 (1.08) | 1e-11 (1.18) |
| Meso. loti | 1e-43 (1.01) | 1e-28 (0.98) | 3e-14 (1.11) | 2e-27 (1.12) | 5e-14 (1.15) |
| Meso. sp. | 3e-43 (1.01) | 1e-22 (0.79) | 4e-10 (1.20) | 5e-30 (1.09) | 5e-13 (1.11) |
| Non-Alpha | Vib. para. 2.1 (1.04) | Symbio. therm. 1.3 (0.89) | Burk. cepacia 0.96 (2.53) | Noc. farcinica 0.17 (1.76) | Pse. syringae 1.7 2.35 |
| Protein | CC1652 [16125898] | CC2247 [16126486] | CC3295 [16127525] | CC1035 [16125287] | |
| Length | 250 | 46 | 169 | 224 | |
| Mag. mag. | 2e-07 (0.91) | 2e-06 (1.61) | 2e-09 (0.52) | - | |
| Rhod. rubr. | 7e-08 (1.01) | - | - | - | |
| Nov. aro. | - | 7e-05 (1.41) | 5e-06 (0.99) | 3e-33 (1.19) | |
| Z. mobilis | - | 1e-04 (1.65) | - | - | |
| C. cres. | 7e-99 (1.00) | 5e-21 (1.00) | 3e-92 (1.00) | e-103 (1.00) | |
| Bra. jap. | 5e-12 (0.87) | 7e-07 (1.63) | 6e-29 (1.00) | 7e-43 (0.90) | |
| Rho. pal. | 5e-12 (0.92) | 5e-06 (2.41) | 2e-27 (1.00) | 3e-42 (0.97) | |
| Agr. tum. | 4e-05 (1.00) | 1e-04 (1.67) | 3e-26 (1.01) | 1e-39 (0.88) | |
| Sino. meli. | 2e-09 (0.89) | 7e-04 (1.67) | 3e-24 (1.16) | 2e-35 (0.89) | |
| Bru. mel. | 2e-11 (0.89) | 0.008 (1.70) | 3e-28 (1.04) | 4e-39 (0.88) | |
| Bru. suis | 1e-11 (0.89) | 0.008 (1.70) | 2e-28 (0.99) | 4e-39 (0.88) | |
| Meso. loti | 2e-06 (0.88) | 5e-05 (1.67) | 9e-24 (1.05) | 1e-38 (0.93) | |
| Meso. sp. | 4e-11 (0.89) | 2e-04 (1.63) | 2e-25 (1.04) | 2e-38 (0.80) | |
| B. henselae | 1e-06 (0.89) | 0.030 (1.70) | 3e-18 (1.02) | 1e-34 (0.88) | |
| B. quintana | 2e-06 (0.89) | 0.002 (1.70) | 8e-18 (1.02) | 2e-35 (0.88) | |
| Non-Alpha | Meth. flagellatus 0.17 (1.53) | - | Burk. cepacia 0.13 (2.89) | Bdell. bacter. 0.70 3.14 | |
a The manner in which these alpha specific proteins were identified is as described in Table 1. Additional abbreviations: Bdell. bacter., Bdellovibrio bacteriovorus; Burk., Burkholderia; Meth., Methylobacillus; Noc., Nocardia; Pse., Pseudomonas; Symbio. therm., Symbiobacterium thermophilum; Vib. para., Vibrio parahaemolyticus.
Table 4.
Signature Proteins Specific for the Rickettsiales or the Rickettsiaceae familya
| Protein | RP104b [15603981] | RP105 [15603982] | RP106c [15603983] | RP766 [15604600] | RP192 [15604066] | RP030 [15603909] | RP187d [15604061] |
| length | 1124 | 672 | 971 | 92 | 128 | 219 | 194 |
| R. prowazekii | 0.0 (1.00) | 0.0 (1.00) | 0.0 (1.00) | 8e-37 (1.00) | 2e-41 (1.00) | e-118 (1.00) | e-108 (1.00) |
| R. conorii | 0.0 (0.88) | 0.0 (0.98) | 0.0 (0.99) | 3e-33 (1.18) | 3e-30 (0.93) | e-107 (1.01) | e-100 (2.56) |
| R. typhi | 0.0 (1.01) | 0.0 (1.00) | 0.0 (1.00) | 1e-27 (0.85) | 3e-40 (1.00) | e-116 (1.00) | e-102 (2.56) |
| R. akari | 0.0 (0.90) | 0.0 (1.00) | 0.0 (1.01) | 2e-25 (0.85) | 2e-35 (1.02) | e-108 (1.01) | 3e-99 (2.56) |
| R. rickettsii | 0.0 (0.88) | 0.0 (0.98) | 0.0 (0.99) | 4e-27 (0.85) | 6e-32 (0.93) | e-109 (1.00) | e-99 (2.56) |
| R. sibirica | 0.0 (0.88) | 0.0 (0.98) | 0.0 (0.99) | 4e-27 (0.85) | 2e-31 (0.93) | e-106 (1.01) | e-101 (2.56) |
| Ehr. canis | 7e-22 (0.75) | 4e-36 (1.25) | 2e-26 (1.49) | - | - | - | - |
| Ehr. rum. | 2e-22 (0.73) | 2e-37 (1.22) | 3e-23 (1.57) | - | - | - | - |
| Ehr. chaf. | 5e-23 (0.73) | 8e-36 (1.23) | - | - | - | - | - |
| Wolbachia | 1e-22 (0.78) | 7e-27 (1.27) | 3e-20 (0.82) | - | - | - | - |
| Ana. mar. | 2e-17 (0.78) | 4e-25 (1.31) | 2e-24 (1.05) | - | - | - | - |
| Non-Rickettsiales | Meso. loti 0.004 (0.32) | Sil. sp. 0.002 (0.53) | Xyl. fast. 2e-07 (0.36) | Leg. pneu. 1.3 (2.60) | M. thermo. 8.1 (2.47) | Myc. pulm. 0.001 (4.72) | Camp. lari 0.69 (4.09) |
a Rickettsiale and Rickettsia specific proteins were identified by whole-genome BLAST searches using protein sequences as probes from the fully sequenced R. prowazekii genome. The RP numbers refer to the protein identification number in the R. prowazekii genome. Other details are as in Table 1 and in Methods section. Additional abbreviations: Camp., Campylobacter; Ehr. chaf., Ehrlichia chaffeensis; Leg. pneu., Legionella pneumophila; M. thermo., Moorella thermoacetica; Myc. pulm., Mycoplasma pulmonis; Xyl. fast., Xylella fastidiosa.
b,cBLAST hits for the family Anaplasmataceae do not show homology over the entire range of the protein and may represent a conserved protein domain.
dBLAST hits for other Rickettsia strains are longer (497 aa) but contain a region that is almost completely homologous to the query sequence.
Table 5.
Signature Proteins Specific for the Rizobiales ordera
| Protein | BQ00140 [49473701] | BQ00720 [49473755] | BQ03880 [49474026] | BQ12030 [49474691] | BQ07670 [49474353] | BQ11900 [49474679] |
| Length | 222 | 83 | 198 | 91 | 336 | 172 |
| Bra. jap. | 5e-18 (1.10) | 2e-09 (1.08) | 2e-20 (0.97) | 2e-07 (1.05) | 5e-46 (1.06) | 1e-17 (0.98) |
| Rho. pal. | 3e-14 (1.11) | 3e-09 (1.06) | 2e-18 (0.97) | 1e-07 (1.37) | - | - |
| Agr. tum. | 5e-13 (1.06) | 6e-11 (1.02) | 1e-26 (0.98) | 2e-12 (1.03) | 7e-66 (0.97) | 2e-24 (1.37) |
| Sino. meli. | 3e-20 (1.00) | 2e-13 (1.02) | 7e-23 (0.98) | 2e-14 (0.98) | 5e-62 (1.02) | - |
| Bru. mel. | 9e-39 (0.98) | 4e-19 (1.04) | 1e-38 (0.97) | 3e-13 (0.54) | 8e-70 (0.96) | 2e-26 (1.02) |
| Bru. suis | 4e-39 (0.98) | 4e-19 (0.96) | 1e-38 (1.03) | 6e-20 (0.99) | 8e-70 (0.98) | 6e-27 (0.98) |
| Meso. loti | 3e-25 (1.07) | 2e-13 (1.18) | 1e-25 (0.98) | 4e-16 (0.99) | 2e-67 (0.95) | 1e-26 (1.03) |
| Meso. sp. | 7e-18 (1.06) | 2e-13 (1.02) | 2e-25 (0.98) | 2e-14 (1.02) | 3e-61 (0.93) | 7e-31 (1.02) |
| B. henselae | 1e-92 (0.97) | 6e-43 (1.00) | 5e-92 (1.00) | 2e-39 (1.00) | e-158 (1.01) | 3e-81 (1.00) |
| B. Quintana | e-127 (1.00) | 4e-43 (1.00) | e-107 (1.00) | 1e-43 (1.00) | 0.0 (1.00) | 1e-91 (1.00) |
| Non – Rizobiale | Bdell. bacter. 0.25 (1.77) | Sil. sp. 0.46 (0.82) | Vibrio fischeri 0.005 (2.49) | St. pyogenes 0.12 (0.86) | St. agalactiae 0.38 (2.65) | Croc. watsonii 6.1 (2.85) |
aSignature proteins that are distinctive of the order Rhizobiales were identified by carrying out BLAST searches of all proteins found in the genome of B. quintana. The BQ numbers refer to the protein identification number in the B. quintana genome. Abbreviations and further details regarding BLAST results are as in Table 1 and the Methods section.. Additional abbreviations: Bdell. bacter., Bdellovibrio bacteriovorus; Croc., Crocosphaera; St., Streptococcus.
Table 6.
Signature Proteins specific for the Rizobiales except the Bradyrhizobiaceae familya
| Protein | BQ01660 [49473833] | BQ02450 [49473907] | BQ03770 [49474017] | BQ13470 [49474819] |
| Length | 119 | 199 | 280 | 179 |
| Bra. jap. | - | - | - | - |
| Rho. pal. | - | - | - | - |
| Agr. tum. | 6e-15 (1.04) | 3e-06 (1.07) | 2e-07 (1.09) | 4e-11 (1.01) |
| Sino. meli. | 1e-15 (1.03) | 1e-05 (1.03) | 1e-11 (1.06) | 1e-10 (1.00) |
| Bru. mel. | 2e-23 (1.06) | 8e-11 (1.01) | 1e-17 (1.07) | 4e-26 (0.99) |
| Bru. suis | 2e-23 (1.06) | 1e-11 (1.12) | 1e-17 (1.21) | 4e-26 (0.99) |
| Meso. loti | 3e-12 (1.39) | 1e-05 (1.25) | 3e-13 (0.96) | 3e-24 (1.00) |
| Meso. sp. | 2e-12 (1.08) | 2e-09 (1.02) | 6e-17 (0.95) | 8e-09 (0.99) |
| B. henselae | 2e-55 (1.00) | 1e-64 (0.99) | 2e-91 (1.01) | 2e-67 (0.99) |
| B. Quintana | 1e-64 (1.00) | e-102 (1.00) | e-131 (1.00) | 2e-99 (1.00) |
| Non – Rizobiale | Bacillus licheniformis 0.77 (1.78) | Treponema denticola 2.9 (2.32) | Therm. tengcongensis 0.001 (2.79) | Mag. mag. 0.60 (1.04) |
See table 1 legend for abbreviations and additional information pertaining to BLAST results. Additional abbreviations: Therm., Thermoanaerobacter.
Table 7.
Signature Proteins specific to the Bartonellaceae family.a
| Protein | BQ12190 [49474706] | BQ11460 [49474647] | BQ11450 [49474646] | BQ11430 [49474645] | BQ11380 [49474640] | BQ11160 [49474626] | BQ11120 [49474623] | BQ11100 [49474621] | BQ11030 [49474614] |
| Length | 94 | 103 | 129 | 65 | 76 | 104 | 264 | 231 | 148 |
| B. henselae | 2e-27 (1.00) | 2e-48 (1.02) | 2e-52 (1.00) | 3e-22 (1.00) | 1e-22 (0.83) | 4e-41 (1.05) | e-103 (1.00) | 2e-94 (1.00) | 3e-64 (0.99) |
| B. quintana | 1e-40 (1.00) | 3e-52 (1.00) | 3e-61 (1.00) | 2e-31 (1.00) | 4e-38 (1.00) | 6e-54 (1.00) | e-145 (1.00) | e-131 (1.00) | 4e-83 (1.00) |
| Other Rizobiales | - | - | - | - | - | - | - | - | - |
| Non-Rizobiale | Prov. rettgeri 1.0 (2.39) | Symbio. therm. 0.024 (2.16) | Bacillus subtilis 3.9 (3.59) | Lacto. gasseri 2.3 (13.8) | Staph. aureus 0.12 (5.84) | Oceano. iheyensis 1.7 (6.16) | Dehalo. etheno. 0.57 (3.08) | Bacillus cereus 1.7 (1.82) | Citro. freundii 1.8 (5.72) |
aAbbreviations and further details regarding BLAST results can be found in table 1. Additional abbreviations: Citro., Citrobacter; Dehalo. etheno., Dehalococcoides ethenogenes; Lacto., Lactobacillus; Oceano., Oceanobacillus; Prov., Providencia; Staph., Staphylococcus; Symbio. therm., Symbiobacterium thermophilum.
Table 8.
Other Alpha-proteobacterial Specific Proteinsa
| Protein | CC0189 [16124444] | CC0569 [16124823] | CC0331 [16124586] |
| Length | 88 | 288 | 186 |
| C. cres. | 5e-38 (1.00) | e-126 (1.00) | e-102 (1.00) |
| Other Alphas |
Mag. mag.; 8e-08 (0.93) Rhod. rubr.; 8e-08 (0.76) Nov. aro.; 1e-10 (0.75) Sil. pom.; 2e-09 (0.67) Sil. sp.; 2-09 (0.68) Rh. spha.; 2e-10 (0.67) |
Nov. aro.; 1e-40 (1.12) Meso. loti; 3e-50 (1.00) |
Bra. jap.; 2e-19 (0.71) Agr. tum.; 2e-23 (0.87) Bru. mel.; 1e-17 (0.76) Bru. suis; 2e-23 (0.92) Meso. loti; 6e-20 (0.89) |
| Non-Alpha | Nitro. euro.; 0.089 (0.84) | Desulf. haf.; 7e-05 (0.88) | Micro. deg.; 0.007 (0.96) |
| Protein | CC0349 [16124604] | CC2323 [16126562] | CC2637 [16126872] |
| Length | 265 | 377 | 374 |
| C. cres. | e-138 (1.00) | 0.0 (1.00) | 0.0 |
| Other Alphas |
Mag. mag.; 2e-29 (0.98) Rhod. rubr.; 8e-32 (0.67) Nov. aro.; 3e-44 (0.92) Sil. sp.; 5e-27 (0.68) |
Mag. mag.; 2e-70 (0.98) Rhod. rubr.; 2e-68 (1.00) Bra. jap.; 3e-40 (1.04) Rho. pal.; 4e-36 (1.03) Agr. tum.; 7e-23 (1.02) Sino. meli.; 3e-27 (1.00) Meso. sp.; 7e-23 (1.01) |
Mag. mag.; 3e-19 (0.99) Rhod. rubr.; 1e-23 (1.09) Bra. jap.; 7e-22 (0.99) Rho. pal.; 8e-19 (0.97) Agr. tum.; 2e-04 (0.91) Sino. meli.; 8e-13 (1.01) Meso. loti; 1e-07 (0.92) Meso. sp.; 1e-06 (1.02) |
| Non-Alpha | Staph. epi.; 0.006 (1.34) | Trep. pallidum; 0.085 (1.14) | Pse. fluor.; 0.32 (1.74) |
a Abbreviations and other details regarding BLAST results can be found in Table 1. Additional abbreviations: Desulf. haf., Desulfitobacterium hafniense; Micro. deg., Microbulbifer degradans; Nitro. euro., Nitrosomonas europaea; Pse. fluor., Pseudomonas fluorescens; Staph. epi., Staphylococcus epidermidis; Trep., Treponema.
Table 9.
Alpha-proteobacterial specific proteins with lateral gene transfers. a
| Protein | CC2585 [16126823] | CC0226 [16124481] |
| Length | 209 | 132 |
| Alpha-Proteobacteria |
Mag. mag.; 2e-16 (0.79) Rhod. rubr.; 2e-16 (1.03) C. cres.; 2e-96 (1.00) Rho. pal.; 7e-21 (1.05) Agr. tum.; 7e-19 (1.02) Sini. mel.; 2e-19 (1.14) Bru. mel.; 5e-19 (1.04) Bru. suis.; 5e-19 (1.04) Meso. loti.; 6e-35 (1.32) |
Rhod. rubr.; 8e-14 (0.59) C. cres.; 6e-72 (1.00) Agr. tum.; 2e-17 (0.64) Sini. mel.; 2e-15 (0.64) Meso. loti.; 1e-18 (0.75) Meso. sp.; 5e-21 (0.64) |
| Other Bacteria | Gamma-proteobacteria: Az. vine.; 2e-07 (1.05) Pse. fluor.; 3e-07 (1.04) Pse. aer.; 1e-06 (1.05) Pse. syr.; 1e-06 (1.04) |
Gamma-proteobacteria: Pse. aer.; 6e-21 (0.64) Sal. ent.; 1e-20 (0.64) |
| Non-Alpha | Des. vulgaris; 0.073 (1.40) | Pro. marinus; 0.26 (2.54) |
| Protein | CC2790 [16127022] | RP382 [15604247] |
| Length | 567 | 510 |
| Alpha-Proteobacteria |
Mag. mag.; 9e-49 (0.41) Nov. aro.; 2e-74 (0.78) C. cres.; 0.0 (1.00) Sil. pom.; 2e-85 (0.72) Sil. sp.; 2e-79 (0.76) Rh. spha.; 1e-79 (0.74) Rho. pal.; 3e-77 (0.68) Agr. tum. 6e-82 (0.72) Bru. mel.; 1e-86 (0.76) Bru. suis;; 6e-83 (0.68) Meso. sp.; 2e-28 (1.40) |
R. prowazekii; 0.0 (1.00) R. conorii; e-155 (0.99) R. akari; e-141 (1.01) R. rickettsii; e-155 (0.99) R. sibirica; e-152 (0.99) Ehr. canis; 9e-27 (0.65) Ehr. rum.; 2e-32 (0.85) Wolbachia; 2e-32 (0.82) Ana. mar.; 6e-20 (0.85) |
| Other Bacteria | Beta-proteobacteria: Burk. cepacia; 4e-18 (0.81) |
Aquificales: Aqu. aeolicus; 7e-11 (0.76) |
| Non-Alpha | Strep. coelicolor; 2.7 (0.65) | Bac. frag.; 0.097 (1.00) |
a Abbreviations and other details regarding BLAST results can be found in Table 1. Additional abbreviations: Az. vine., Azotobacter vinelandii; Aqu., Aquifex; Bac. frag., Bacteroides fragilis; Burk. Burkholderia; Des., Desulfovibrio; Lep. int., Leptospira interrogans; Pro., Prochlorococcus; Pse. aer., Pseudomonas aeruginosa; Pse. fluor., Pse. fluorescens; Pse. syr., Pse. syringae; Sal. ent., Salmonella enterica.
The first grouping of α-proteobacterial markers consists of 6 proteins that are specific to nearly all sequenced α-proteobacterial species and are not found in any other Bacteria (Table 1). These proteins clearly distinguish the α-proteobacteria as a distinct group from all other Bacteria. Even though some genes have been lost from certain species, these proteins remain largely distinctive of the α-subdivision. Interestingly, no homologues were detected in Zymomonas mobilis for three of these signature proteins (CC3319, CC1887, CC1725). Z. mobilis is also lacking a number of other signature proteins described in this study and this may be attributed to the genetic loss of a variety genes resulting in its small genome size (2.06 Mb) [33]. A number of genes for the tricarboxcylic acid cycle as well as other functions have previously been documented as missing in this genome [33]. One of these signature proteins (CC1725) is also not found in Novosphingobium aromaticivorans indicating it was lost from members of the Sphingomonadales family. A homologue of the protein CC3319 was detected in the currently unclassified Magnetococcus sp. MC-1 genome suggesting that this species may be distantly related to the α-proteobacteria [42]. A number of α-proteobacteria-specific indels (i.e., inserts or deletions) are also present in Magnetococcus [17], supporting the above inference. Finally, the protein CC1887 is also found in the α-proteobacteria as well as a variety of Eukaryotes supporting the derivation of mitochondrion from an α-proteobacterial lineage [9-13].
Another group of 10 signature proteins showing a high affinity for sequenced alphas are those distinguishing all other α-proteobacteria from the order Rickettsiales (Table 2). In this case, the Rickettsiales show no detectable homologues of otherwise α-specific proteins. These results suggests that the genes for these proteins have either been lost from the Rickettsiales or it forms one of the earliest branching lineage within α-proteobacteria [2,43]. These proteins are present in almost all other sequenced α-proteobacteria with few exceptions. The proteins CC0520 and CC0366 have homologues in Magnetococcus sp. MC-1 again lending support to the inference that this unclassified species is distantly related to the alpha-group. The protein CC1977 is also found in Eukaryotes and the E values for a few representative eukaryotic species are given in the Table 2 legend. One protein (CC3010), showing a very high affinity for this grouping as noted by low E values, is also found in a single gamma proteobacterium (Pseudomonas sp.). This finding is most likely due to a non-specific event such as a lateral gene transfer (LGT) of which additional examples will be presented later.
The next grouping of signature proteins are those which are found in almost all sequenced α-proteobacteria excluding the intracellular pathogens belonging to the Bartonellaceae family and the order Rickettsiales (Table 3A). This grouping outlines a case in which proteins have probably been lost independently from two unrelated groups within the α-proteobacteria most likely due to their intracellular lifestyles [2,3,44]. Five proteins of this type were identified with minimal loses seen in other α-proteobacteria. CC2345 provides a good example of this type of protein since it is highly conserved in all available α-proteobacterial genomes as indicated by low E values. The other four proteins also show a high affinity for this category with losses occurring only in Z. mobilis and Rhodospirillum rubrum.
A variation on the above theme is a collection of 4 α-specific proteins that are absent in the orders Rickettsiales and Rhodobacterales (Table 3B). However, a key feature distinguishing these proteins from those presented in Table 3A is the free-living lifestyle of the Rhodobacterales as opposed to the intracellular Bartonellas. Since Rickettsiales and the Rhodobacterales are not known to share any unique characteristic, it is possible that the loss of these proteins from these two orders has occurred due to unrelated reasons. Also, some additional losses are seen in this grouping. For example the protein CC1652 is absent in the Sphingomonadales while the protein CC1035 is absent in the Rhodospirillales. Note that the protein CC2247 exhibits high E values for BLAST hits representing Brucellaceae and Bartonellaceae but this high E value is acceptable due to the very short length of this protein (46 amino acids) and the fact that besides α-proteobacteria no other BLAST hits were observed (Table 3B).
The Blast searches on proteins found in the R. prowazekii genome have led to identification of a number of signature proteins which are specific to species belonging to the order Rickettsiales. This order is made up of two families: the Anaplasmataceae (Anaplasma, Ehrlichia and Wolbachia) and Rickettsiaceae (Rickettsias) [2,43]. The first group of such proteins (RP104, RP105, and RP106) are present in all species belonging to the order Rickettsiales, but are not found in any other α-proteobacteria (Table 4). It should be noted that the proteins RP104 and RP106 do not show homology over the entire length of the homologous proteins in members of the Anaplasmataceae family. Thus, additional domains that are specific for the Rickettsiaceae family may be present in these proteins. These signature proteins are highly conserved within this order, as indicated by their very low E values (Table 4) and represent interesting examples of genes that were likely introduced in a common ancestor of the Rickettsiales. Note that the first non-Rickettsiale BLAST hit for the protein RP106 appears at 2e-07 (Xyella fastidiosa). RP106 is still included as a Rickettsiales-specific protein because the Xyella protein is only 348 amino acids in length and thus it is likely a different protein.
Another group of 4 proteins are specific to the Rickettsia species and are not found in other members of the Rickettsiales (Table 4). These proteins (RP766, RP192, RP030 and RP187) are highly conserved and represent cases in which genes were introduced into a common ancestor of the Rickettsiaceae. Homologues of the protein RP187 are much longer in other Rickettsia strains (194 vs 497 aa) but the region representing the query sequence is highly conserved. It is possible that other Rickettsia species have acquired an additional protein domain during the course of evolution.
In addition to the Rickettsiales, the Rhizobiales form a major order within the α-proteobacteria [1,17,42]. To identify proteins which are distinctive of the Rhizobiales, BLAST searches were carried out on all ORFs in the genome of B. quintana. Six proteins have been identified that are conserved amongst all sequenced Rhizobiales with minimal evidence of gene loss occurring (Table 5). The protein BQ07670 is absent in Rhodopseudomonas palustris while the protein BQ11900 is absent in this strain as well as in Sinorhizobium meliloti. The presence of these proteins solely in the Rhizobiales indicates they were likely introduced in a common ancestor of this order.
Other signature proteins that are useful in defining the Rhizobiales are those which are present in all sequences members of this order, except the Bradyrhizobiaceae family (Table 6). Four proteins of this type have been identified with no losses occurring in any species. These proteins indicate that the Bradyrhizobiaceae family is more distantly related to other members of the Rhizobiales. The deeper branching and distinctness of Bradyrhizobiaceae and Methylobacteriaceae from other Rhizobiales is also strongly supported by phylogenetic analyses based on different gene sequences and conserved indels in many proteins [1,17,45].
A number of proteins have also been identified which are unique to the Bartonella species. Nine examples of such proteins are shown in Table 7. These proteins are highly conserved amongst both sequenced Bartonella species with no gene losses occurring. The presence of these proteins solely in this family of α-proteobacteria indicates that they should provide useful markers for the Bartonellaceae family.
Six other α-specific signature proteins were identified that do not show any distinct pattern but are sporadically present in α-proteobacterial species (Table 8). These proteins are more randomly distributed among a limited number of sequenced α-proteobacteria and it is likely that gene losses for these proteins have occurred independently in various species or groups. Nevertheless, these proteins are still unique to the α-proteobacteria. The protein CC0189 is represented in the Rhodospirillales, Novosphingomonadales, Caulobacterales and Rhodobacterales but is not found in any Rhizobiales. One protein (CC0331) is represented in various families within the Rhizobiales while two others (CC2323 and CC2637) show a similar trend and are also present in Rhodospirillales.
A final grouping of 4 signature proteins consists of those where limited lateral gene transfers (LGTs) have apparently occurred (Table 9). Three of these proteins (CC2585, CC0226 and CC2790) were isolated from the Caulobacter genome and represent cases in which genes were also present in a limited numbers of gamma or beta-proteobacteria. Specifically, a homologue of the protein CC2585 was detected in a number of gamma-proteobacteria belonging to the Pseudomonadaceae family while CC0226 was only detected in Pse. aeruginosa and the enteric bacterium Salmonella enterica. The protein CC2790 shows some similarity to a Superfamily I DNA and RNA helicase found in Burkholderia cepacia (beta-proteobacteria). However, this BLAST hit only shows conservation over 142 amino acids of the 567 amino acids C. crescentus protein. Furthermore, all alpha BLAST hits are annotated as hypothetical proteins indicating this non-alpha BLAST hit probably represents a different protein with a shared protein domain that was transferred. Interestingly, one of the proteins, RP382, which is otherwise highly specific for the order Rickettsiales, is also found in Aquifex aeolicus. In each of these cases, the direction of gene transfer remains unclear.
Discussion
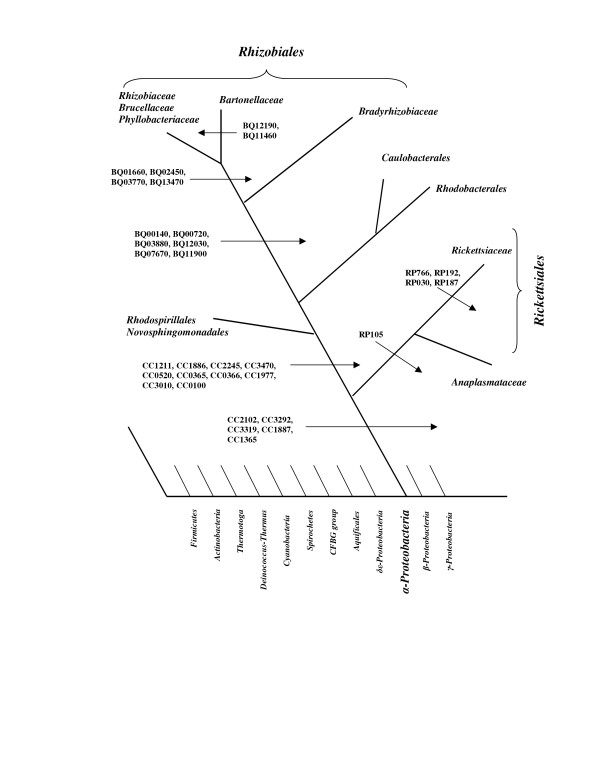
The α-proteobacteria forms an extremely diverse group showing vast differences in such characteristics as morphology, metabolism, and physiology [1]. In the current view, this group is distinguished from all other Bacteria based on 16S rRNA phylogenetic trees [1,8,19,46]. Few molecular or physiological characteristics were known which clearly distinguish this group from all other Bacteria [1,7]. However, our recent work has identified a large number of conserved inserts and deletions in protein sequences which are distinctive characteristics of α-proteobacteria and its subgroups and not found in any other groups of Bacteria [17] (see also http://www.bacterialphylogeny.com). These signatures provide useful tools for identifying α-proteobacteria within Bacteria as well as for understanding the interrelationships and branching order within this group. Here, we describe 61 signature proteins that are largely specific for the α-proteobacteria. Almost all of these proteins are of hypothetical functions, and in view of their α-proteobacterial specificity, it is likely that they are involved in functions that are limited to only this group of bacteria. Because such genes are likely involved in specialized functions, the loss of some of these genes from certain α-proteobacterial species is not surprising. Based on signature proteins described here, along with various α-proteobacteria-specific conserved inserts and deletions [17], a clearer picture of α-proteobacteria phylogeny and taxonomic classification can be derived. Figure 1 presents a model for α-proteobacterial evolution which indicates the evolutionary stages where these proteins are suggested to have evolved or been introduced. The model based on these signature proteins is identical to that deduced independently based upon a large number of conserved indels in different proteins [17], indicating its reliability.
Figure 1.
Summary diagram showing the distribution pattern of various α-proteobacteria signature proteins. The arrows indicate the evolutionary stages where these signature proteins were likely introduced. Some proteins, which are sporadically present in α-proteobacteria are not shown here. The branching position of α-proteobacteria relative to other bacterial groups was deduced as described in earlier work [13,17,40].
Several signature proteins are specific to nearly all α-proteobacteria. These proteins provide additional support to various alpha-distinguishing indels, which are found only in the α-proteobacteria and not in any other groups of bacteria. Examples of such indels include the following: an 8 amino acid insert in the α subunit of ATP synthase complex, 3 amino acid insert in prolipoprotein-phosphatidylglycerol transferase, and a 1 amino acid deletion in the FtsK protein [17]. The simplest and most parsimonious explanation for the presence of these α-specific signatures (both proteins and indels) is that they were introduced once in a common ancestor of all α-proteobacteria and their presence in various α-proteobacterial species is due to vertical transmission [47,48]. It is difficult to explain the presence of these genes in various α-proteobacteria by other non-specific means such as lateral gene transfers [49]. The finding of these unique genes and conserved indels in various α-subdivision members strongly indicates that all such bacteria carry out certain physiological functions that are unique to the members of this group. Therefore, studies aimed at determining the functional roles of these proteins and indels are of much interest.
The largest group of signature proteins discovered are those found in all α-proteobacteria excluding the order Rickettsiales. These proteins indicate that the Rickettsiales constitute a distinct clade within the α-subdivision, which is in accordance with phylogenetic analyses based on different gene sequences [2,17,43,50]. Phylogenetic studies based on 16S rRNA and many other genes [2,43,45,50], as well as our studies based on conserved indels in several proteins that are present in various α-proteobacteria but absent in Rickettsiales as well as other groups of bacteria [17], provide evidence that the Rickettsiales comprise the deepest branching group within α-proteobacteria. In view of this, the most logical explanation for these signatures is that they were introduced in a common ancestor of other α-proteobacteria after the divergence of the Rickettsiales (Figure 1).
An interesting group of α-specific signature proteins are those which are absent in the intracellular pathogens belonging to the order Rickettsiales and the family Bartonellaceae. The latter group of species form a family within the Rhizobiales order [1,17]. Because these two groups are phylogenetically unrelated, it is likely that the genes for these proteins were selectively lost in these two groups independently due to their intracellular lifestyles. It is logical to assume that the cellular functions of these proteins are either not required in the intracellular environment, or they are provided for by the host cells leading to the loss of these genes from these organisms. These proteins could have been introduced in either a common ancestor of all α-proteobacteria and subsequently lost in the Rickettsiales and Bartonellaceae, or introduced after the divergence of the Rickettsiales and lost in the Bartonellaceae. It is interesting that the Brucellas (also intracellular pathogens) have retained all of these proteins indicating that this group differ in its physiological requirements from other α-proteobacterial intracellular pathogens [1,3,51]. Several α-specific signature proteins that are absent in both the Rickettsiales as well as Rhodobacterales were also identified. Since there is no evidence to suggest any sort of relationship between these two groups [1,17], the simplest explanation is that these genes were introduced after the divergence of the Rickettsiales and lost preferentially by the Rhodobacterales.
Other signature proteins were isolated pointing to a variety of relationships. For instance, the protein CC0189 which is only present in Caulobacterales, Rhodobacteriales, Rhodospirillales and Novosphingomonadales indicates a close relationship between these deep branching orders within α-proteobacteria. This relationship is also seen from the protein CC0349 but to a lesser extent since losses have occurred in some species. These findings are supported by indels in a variety of proteins that indicate these orders show a closer relationship and have branched prior to the Rhizobiales [17]. Other signature proteins are found in a selection of these above orders and are also found in some but not all families within the Rhizobiales (CC0331, CC2323 and CC2637). A close relationship between Caulobacter and Rhodobacterales is generally indicated by phylogenetic trees and is also supported by a conserved 11 amino acid insert in the protein aspargine-glutamine amido transferase [1,17]. Thus, it is somewhat surprising that in our analysis of the Caulobacter genome, we did not identify any signature protein that was uniquely shared by these two α-proteobacterial orders. However, a 12 amino acid insert in the protein DNA ligase indicates that Rhodobacterales may be more closely related to Rhizobiales in comparison to Caulobacterales [17]. In view of these results, and the fact that C. crescentus represents the only fully sequenced bacterium within its order [24], additional sequence information is required to further clarify the evolutionary relationships amongst Rhizobiales, Rhodobacterales and Caulobacterales.
Several signature proteins were found to be specific for either the order Rickettsiales or the family Rickettsiaceae. These proteins provide molecular markers for these groups and they likely originated in common ancestors of these groups. The distinctness of these groups is also supported by a number of conserved indels in different proteins which are uniquely present in the species from these groups, but not found in any other bacteria [17]. It should be noted that McLeod et al. [28] based upon their comparative analysis of the Rickettsias genomes have identified a number of hypothetical proteins that are only found in particular Rickettsias. These proteins were grouped into the following classes: R. typhi ORFs not found in R. conorii or R. prowazekii; R. typhi ORFs found in R. conorii but not in R. prowazekii; and R. typhi ORFs found in R. prowazekii but not in R. conorii. However, no proteins that were specific for all Rickettsias or Rickettsiales were described in the McLeod et al. study [28].
A number of signature proteins identified here are useful in defining and characterizing the Rhizobiales order. Of the six Rhizobiales-specific proteins described here, four (viz. BQ00140, BQ00720, BQ03880 and BQ12030) are completely conserved amongst all sequenced Rhizobiales and should provide good molecular markers for this order. Two other proteins (BQ07670 and BQ11900) also show a high affinity for this grouping with a few gene losses in some species. We have previously described a conserved indel in tryptophanyl-tRNA synthetase that is present in all sequenced Rhizobiales but is absent in all other bacteria [17]. These signatures were likely introduced in a common ancestor of the Rhizobiales order (Figure 1). Four additional proteins that were identified here are completely specific to all sequenced Rhizobiales, except for the Bradyrhizobiaceae family. Phylogenetic analysis based on a number of gene sequences as well as conserved indels in a number of proteins (viz. Trp-tRNA synthetase, LytB metalloproteinase) provide evidence that that the Bradyrhizobiaceae family is distantly related to other Rhizobiales (Rhizobiaceae, Brucellaceae, Phyllobacteriaceae), and it has branched prior to the latter groups of species [1,17,45]. Thus, it is likely that these signature proteins evolved in a common ancestor of various other Rhizobiales after the divergence of the Bradyrhizobiaceae family (Figure 1). A number of signature proteins that are unique for the Bartonella species were later introduced in that particular branch of the tree (Figure 1).
Although most of the signature proteins identified here are specific for only the α-proteobacteria, we have also come across a few examples where lateral gene transfer seems to have occurred between α-proteobacteria and a few species from other groups of bacteria. The rarity of such proteins in comparison to those which exhibit strict group-specificity indicates that most newly acquired alpha-specific genes have been predominantly transmitted via vertical descent and LGT and other non-specific mechanisms play relatively minor role in their transmission. It should be mentioned that although our analyses of proteins in R. prowazekii, C. crescentus and B. quintana genomes have identified a large number of signature proteins, based on these studies signature proteins for certain other groups within α-proteobacteria (e.g. Rhizobiaceae, Rhodobacterales, Sphignomonadales, etc.) will not be detected. It should be possible to identify signatures for these groups by carrying out similar analysis using protein sequences from these genomes.
Daubin and Ochmann [52] and Lerat et al. [36,47] have previously examined the gene repertoire of γ-proteobacteria and have indicated the presence of many ORFans genes (i.e. ORFs that have no known homologs) that are limited to either certain bacterial strains or particular subgroups of γ-proteobacteria. The ORFan genes were found to be present in their studies in different monophyletic clades at different phylogenetic depths, which is similar to what we have reported here for the signature proteins in the α-proteobacteria taxon. The other characteristics of ORFans genes noted by these authors were that they are generally short (between 400–500 bp), A+T rich, and evolve faster than other genes which are more broadly distributed [47,52]. Many of the signature proteins identified in the present work are of similar lengths as the ORFans genes. These earlier studies also indicate that ORFans genes generally encode for functional proteins, and once acquired they are vertically transmitted, and based on them it possible to make robust phylogenetic inference as we have been able to do in the present study for α-proteobacteria. Although the source of ORFans genes in different genomes remains to be determined, it has been suggested that many of them are derived from bacteriophages [47,52].
The concept that mitochondria have originated from an α-proteobacterial ancestor is supported by a large body of evidence including phylogenetic analysis and shared presence of many common indels [9-14]. The homologues of two of the α-proteobacterial signature proteins (CC1887 and CC1977) are also present in Eukaryotes providing further support for this inference. For the remainder of the proteins no eukaryotic homologues were detected which supports the observation of Boussau et al. [44] that for a large fraction of genes in α-proteobacterial genome no homologs are found in the eukaryotes. Currently, it is thought that within α-proteobacteria the species belonging to the order Rickettsiales are the closest relatives of mitochondria [10-12,53-55]. However, of the two proteins which are commonly found in eukaryotes, only one of them (CC1887) is present in the Rickettsiales. A specific relationship of mitochondria to the Rickettsiales is also supported by only some conserved indels, but not all [17]. In a recent study, where the relationship of alpha proteobacteria to mitochondria was examined based on a large number of individual and concatenated protein sequences [56], the closest relationship of mitochondria was seen for Rhodospirillum rubrum rather than the Rickettsiales. In earlier work, we have described two conserved signatures (a 37 aa insert in valyl-tRNA synthetase and 1 aa indel in LonA protein), which were commonly shared by all eukaryotic homologs and certain other groups of bacteria but which were not found in any α-proteobacteria [13]. An update of these signatures indicates that they still constitute exceptions to the α-proteobacterial derivation of the mitochondrial/eukaryotic homologs (R.S. Gupta, unpublished results). These observations in conjunction with the recent conflicting observations regarding the possible origins of NADH dehydrogenase subunits from Trichomonas vaginalis [57,58] indicate that additional work is necessary to clarify the sources of different mitochondrial and nuclear cytosolic genes of eukaryotic proteins.
Conclusion
Whole-genome analyses of B. quintana, Ri prowazekii and C. crescentus proteins have led to the discovery of 61 signature proteins which are distinctive characteristics of the α-proteobacteria and its subgroups. These signature proteins provide additional support to our recent work based on a large number of conserved inserts and deletions in protein sequences that are either specific for the α-proteobacteria or provide information regarding the interrelationships and branching order within this group [17]. Sequence information from additional α-proteobacterial species will be useful in testing the predicted presence or absence of various identified molecular signatures (indels and proteins) in different groups, thus validating the suggested relationships. Studies aimed at understanding the cellular functions of these α-specific signature proteins should be of much interest since they will likely provide novel insights into unique physiological characteristics shared by this important group of bacteria and its various subgroups. Studies on proteins which are specific to the intracellular pathogens, such as Rickettsiales and Bartonella, could also provide new drug targets for their associated diseases.
Methods
Identification of α-Proteobacteria Specific Proteins
To identify signature proteins which are specific to the α-proteobacteria or its various subgroups, all proteins in the genomes of C. crescentus, R. prowazekii, and B. quintana were analyzed. BLAST searches were carried out [41] on each individual protein in these genomes to identify all other bacteria containing proteins with similar sequences. These results were visually inspected for homologues showing specificity to α-proteobacteria with no other similar homologues present in any other Bacteria. Expect values (E values) were analyzed for putative α-specific proteins. The E values, which are calculated by the BLAST software, indicate the probability that the observed similarity between the query protein and any other protein detected by the BLAST search arose by chance [41]. In BLAST searches, the E values are lowest (closer to 0) for BLAST hits with a high degree of homology to the query sequence and they increase as BLAST hits are detected with lower similarity. The results of BLAST searches were inspected for sudden increases in E values from the last α-proteobacteria in the search to the first non-alpha bacteria. This increase in E values was important when the next non-alpha BLAST hit was in a range where the observed similarity could occur by chance (> 10-05). However, higher E values were sometimes allowed and could be significant for smaller proteins since they contain fewer characters resulting in higher E values (for statistical reasons) for their true homologs. For all α-specific signature proteins described here, E values were recorded for each blast hit as well as for the first non-alpha bacterium in a given search. Although E values take into account the length of the sequence over which the similarity is observed between the query sequence and a BLAST hit, low E values can sometime result if high degree of homology is observed between two different proteins over a short sequence region. Therefore, we have also inspected BLAST results for homology over the entire protein length and for similarity in protein length. The length ratios of the hit proteins over the query protein are shown in brackets beside the E values and these values are expected to be close to 1.00 if the identified proteins are of similar lengths as the query protein. It should be mentioned that BLAST searches can sometime indicate misleading similarity, particularly when no close relatives of the query species are in the database [59]. However, in the present study where most of the BLAST hits correspond to α-proteobacteria, such a possibility is highly unlikely. All proteins indicated in the Tables 1, 2, 3, 4, 5, 6, 7, 8, 9 are specific for the α-proteobacteria based on these criteria unless otherwise mentioned.
Authors' contributions
PK carried out BLAST searches on different proteins and was responsible for the initial evaluation of the results. RSG conceived and directed this study and was responsible for the final evaluation of the results. PK prepared a rough draft of the manuscript under RSG's directions, which was revised and modified by RSG. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
The work was supported by a research grant from the National Science and Engineering Research Council of Canada.
Contributor Information
Pinay Kainth, Email: kainthps@muss.cis.mcmaster.ca.
Radhey S Gupta, Email: gupta@mcmaster.ca.
References
- Kersters K, Devos P, Gillis M, Vandamme P, Stackebrandt E. Introduction to the Proteobacteria. In: Dworkin M, editor. The Prokaryotes: An Evolving Electronic Resource for the Microbiological Community. 3. New York, Springer-Verlag; 2003. Release 3.12 http://141.150.157.117:8080/prokPUB/chaprender/jsp/showchap.jsp?chapnum=379. [Google Scholar]
- Yu XJ, Walker DH. The Order Rickettsiales. In: Dworkin M, editor. The Prokaryotes: An Evolving Electronic Resource for the Microbiological Community. 3. New York, Springer-Verlag; 2003. Release 3.12 http://link.springer-ny.com/link/service/books/10125/ [Google Scholar]
- Moreno E, Moriyon I. The Genus Brucella. In: Dworkin M, editor. The Prokaryotes: An Evolving Electronic Resource for the Microbiological Community. 3. New York, Springer-Verlag; 2001. Release 3.7 http://link.springer-ny.com/link/service/books/10125/ [Google Scholar]
- Sadowsky MJ, Graham PH. Root and Stem Nodule Bacteria of Legumes. In: Dworkin M, editor. The Prokaryotes: An Evolving Electronic Resource for the Microbiological Community. 3. New York, Springer-Verlag; 2000. Release 3.3 http://link.springer-ny.com/link/service/books/10125/ [Google Scholar]
- Sawada H, Kuykendall LD, Young JM. Changing concepts in the systematics of bacterial nitrogen-fixing legume symbionts. J Gen Appl Microbiol. 2003;49:155–179. doi: 10.2323/jgam.49.155. [DOI] [PubMed] [Google Scholar]
- Van Sluys MA, Monteiro-Vitorello CB, Camargo LE, Menck CF, da Silva AC, Ferro JA, Oliveira MC, Setubal JC, Kitajima JP, Simpson AJ. Comparative genomic analysis of plant-associated bacteria. Annu Rev Phytopathol. 2002;40:169–189. doi: 10.1146/annurev.phyto.40.030402.090559. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E, Murray RGE, Trüper HG. Proteobacteria classis nov., a name for the phylogenetic taxon that includes the "Purple bacteria and their Relatives". Int J Syst Bacteriol. 1988;38:321–325. [Google Scholar]
- De Ley J. The Proteobacteria: Ribosomal RNA cistron similarities and bacterial taxonomy. In: Balows A, Trüper HG, Dworkin M, Harder W and Schleifer KH, editor. The Prokaryotes. 2. Vol. 100. New York, Springer-Verlag; 1992. pp. 2111–2140. [Google Scholar]
- Margulis L. Origin of Eukaryotic cells. New Haven, CT., Yale University Press; 1970. [Google Scholar]
- Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Ponten T, Alsmark UC, Podowski RM, Naslund AK, Eriksson AS, Winkler HH, Kurland CG. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- Emelyanov VV. Evolutionary relationship of Rickettsiae and mitochondria. FEBS Letters. 2001;501:11–18. doi: 10.1016/S0014-5793(01)02618-7. [DOI] [PubMed] [Google Scholar]
- Gupta RS. The phylogeny of Proteobacteria: relationships to other eubacterial phyla and eukaryotes. FEMS Microbiol Rev. 2000;24:367–402. doi: 10.1016/S0168-6445(00)00031-0. [DOI] [PubMed] [Google Scholar]
- Viale AM, Arakaki AK. The chaperone connection to the origins of the eukaryotic organelles. FEBS Letters. 1994;341:146–151. doi: 10.1016/0014-5793(94)80446-X. [DOI] [PubMed] [Google Scholar]
- Gupta RS, Golding GB. The origin of the eukaryotic cell. Trends Biochem Sci. 1996;21:166–171. doi: 10.1016/0968-0004(96)20013-1. [DOI] [PubMed] [Google Scholar]
- Gupta RS. Protein Phylogenies and Signature Sequences: A Reappraisal of Evolutionary Relationships Among Archaebacteria, Eubacteria, and Eukaryotes. Microbiol Mol Biol Rev. 1998;62:1435–1491. doi: 10.1128/mmbr.62.4.1435-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RS. Protein signatures distinctive of Alpha proteobacteria and its subgroups and a model for Alpha proteobacterial evolution. Crit Rev Microbiol. 2005;31:101–135. doi: 10.1080/10408410590922393. [DOI] [PubMed] [Google Scholar]
- Olsen GJ, Woese CR, Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994;176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Stackebrandt E, Weisburg WG, Paster BJ, Madigan MT, Fowler CMR, Hahn CM, Blanz P, Gupta R, Nealson KH, Fox GE. The phylogeny of purple bacteria: the alpha subdivision. System Appl Microbiol. 1984;5:315–326. doi: 10.1016/s0723-2020(84)80034-x. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Nakamura Y, Sato S, Asamizu E, Kato T, Sasamoto S, Watanabe A, Idesawa K, Ishikawa A, Kawashima K, Kimura T, Kishida Y, Kiyokawa C, Kohara M, Matsumoto M, Matsuno A, Mochizuki Y, Nakayama S, Nakazaki N, Shimpo S, Sugimoto M, Takeuchi C, Yamada M, Tabata S. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 2000;7:331–338. doi: 10.1093/dnares/7.6.331. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Nakamura Y, Sato S, Minamisawa K, Uchiumi T, Sasamoto S, Watanabe A, Idesawa K, Iriguchi M, Kawashima K, Kohara M, Matsumoto M, Shimpo S, Tsuruoka H, Wada T, Yamada M, Tabata S. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 2002;9:189–197. doi: 10.1093/dnares/9.6.189. [DOI] [PubMed] [Google Scholar]
- Wood DW, Setubal JC, Kaul R, Monks DE, Kitajima JP, Okura VK, Zhou Y, Chen L, Wood GE, Almeida NFJ, Woo L, Chen Y, Paulsen IT, Eisen JA, Karp PD, Bovee DS, Chapman P, Clendenning J, Deatherage G, Gillet W, Grant C, Kutyavin T, Levy R, Li MJ, McClelland E, Palmieri A, Raymond C, Rouse G, Saenphimmachak C, Wu Z, Romero P, Gordon D, Zhang S, Yoo H, Tao Y, Biddle P, Jung M, Krespan W, Perry M, Gordon-Kamm B, Liao L, Kim S, Hendrick C, Zhao ZY, Dolan M, Chumley F, Tingey SV, Tomb JF, Gordon MP, Olson MV, Nester EW. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science. 2001;294:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- DelVecchio VG, Kapatral V, Redkar RJ, Patra G, Mujer C, Los T, Ivanova N, Anderson I, Bhattacharyya A, Lykidis A, Reznik G, Jablonski L, Larsen N, D'Souza M, Bernal A, Mazur M, Goltsman E, Selkov E, Elzer PH, Hagius S, O'Callaghan D, Letesson JJ, Haselkorn R, Kyrpides N. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc Natl Acad Sci USA. 2002;99:443–448. doi: 10.1073/pnas.221575398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierman WC, Feldblyum TV, Laub MT, Paulsen IT, Nelson KE, Eisen J, Heidelberg JF, Alley MR, Ohta N, Maddock JR, Potocka I, Nelson WC, Newton A, Stephens C, Phadke ND, Ely B, DeBoy RT, Dodson RJ, Durkin AS, Gwinn ML, Haft DH, Kolonay JF, Smit J, Craven MB, Khouri H, Shetty J, Berry K, Utterback T, Tran K, Wolf A, Vamathevan J, Ermolaeva M, White O, Salzberg SL, Venter JC, Shapiro L, Fraser CM. Complete genome sequence of Caulobacter crescentus. Proc Natl Acad Sci U S A. 2001;98:4136–4141. doi: 10.1073/pnas.061029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen IT, Seshadri R, Nelson KE, Eisen JA, Heidelberg JF, Read TD, Dodson RJ, Umayam L, Brinkac LM, Beanan MJ, Daugherty SC, DeBoy RT, Durkin AS, Kolonay JF, Madupu R, Nelson WC, Ayodeji B, Kraul M, Shetty J, Malek J, Van Aken SE, Riedmuller S, Tettelin H, Gill SR, White O, Salzberg SL, Hoover DL, Lindler LE, Halling SM, Boyle SM, Fraser CM. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc Natl Acad Sci U S A. 2002;99:13148–13153. doi: 10.1073/pnas.192319099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larimer FW, Chain P, Hauser L, Lamerdin J, Malfatti S, Do L, Land ML, Pelletier DA, Beatty JT, Lang AS, Tabita FR, Gibson JL, Hanson TE, Bobst C, Torres JL, Peres C, Harrison FH, Gibson J, Harwood CS. Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat Biotechnol. 2004;22:55–61. doi: 10.1038/nbt923. [DOI] [PubMed] [Google Scholar]
- Ogata H, Audic S, Renesto-Audiffren P, Fournier PE, Barbe V, Samson D, Roux V, Cossart P, Weissenbach J, Claverie JM, Raoult D. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science. 2001;293:2093–2098. doi: 10.1126/science.1061471. [DOI] [PubMed] [Google Scholar]
- McLeod MP, Qin X, Karpathy SE, Gioia J, Highlander SK, Fox GE, McNeill TZ, Jiang H, Muzny D, Jacob LS, Hawes AC, Sodergren E, Gill R, Hume J, Morgan M, Fan G, Amin AG, Gibbs RA, Hong C, Yu XJ, Walker DH, Weinstock GM. Complete genome sequence of Rickettsia typhi and comparison with sequences of other rickettsiae. J Bacteriol. 2004;186:5842–5855. doi: 10.1128/JB.186.17.5842-5855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galibert F, Finan TM, Long SR, Puhler A, Abola P, Ampe F, Barloy-Hubler F, Barnett MJ, Becker A, Boistard P, Bothe G, Boutry M, Bowser L, Buhrmester J, Cadieu E, Capela D, Chain P, Cowie A, Davis RW, Dreano S, Federspiel NA, Fisher RF, Gloux S, Godrie T, Goffeau A, Golding B, Gouzy J, Gurjal M, Hernandez-Lucas I, Hong A, Huizar L, Hyman RW, Jones T, Kahn D, Kahn ML, Kalman S, Keating DH, Kiss E, Komp C, Lelaure V, Masuy D, Palm C, Peck MC, Pohl TM, Portetelle D, Purnelle B, Ramsperger U, Surzycki R, Thebault P, Vandenbol M, Vorholter FJ, Weidner S, Wells DH, Wong K, Yeh KC, Batut J. The composite genome of the legume symbiont Sinorhizobium meliloti. Science. 2001;293:668–672. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- Brayton KA, Kappmeyer LS, Herndon DR, Dark MJ, Tibbals DL, Palmer GH, McGuire TC, Knowles DPJ. Complete genome sequencing of Anaplasma marginale reveals that the surface is skewed to two superfamilies of outer membrane proteins. Proc Natl Acad Sci U S A. 2005;102:844–849. doi: 10.1073/pnas.0406656102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins NE, Liebenberg J, de Villiers EP, Brayton KA, Louw E, Pretorius A, Faber FE, van Heerden H, Josemans A, van Kleef M, Steyn HC, van Strijp MF, Zweygarth E, Jongejan F, Maillard JC, Berthier D, Botha M, Joubert F, Corton CH, Thomson NR, Allsopp MT, Allsopp BA. The genome of the heartwater agent Ehrlichia ruminantium contains multiple tandem repeats of actively variable copy number. Proc Natl Acad Sci U S A. 2005;102:838–843. doi: 10.1073/pnas.0406633102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsmark CM, Frank AC, Karlberg EO, Legault BA, Ardell DH, Canback B, Eriksson AS, Naslund AK, Handley SA, Huvet M, La Scola B, Holmberg M, Andersson SG. The louse-borne human pathogen Bartonella quintana is a genomic derivative of the zoonotic agent Bartonella henselae. Proc Natl Acad Sci U S A. 2004;101:9716–9721. doi: 10.1073/pnas.0305659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo JS, Chong H, Park HS, Yoon KO, Jung C, Kim JJ, Hong JH, Kim H, Kim JH, Kil JI, Park CJ, Oh HM, Lee JS, Jin SJ, Um HW, Lee HJ, Oh SJ, Kim JY, Kang HL, Lee SY, Lee KJ, Kang HS. The genome sequence of the ethanologenic bacterium Zymomonas mobilis ZM4. Nat Biotechnol. 2005;23:63–68. doi: 10.1038/nbt1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone DR, Castenholz RW, Garrity GM. Bergey's Manual of Systematic Bacteriology. 2nd. One, The Archaea and the Deeply branching and phototrophic bacteria. New York, Springer; 2001. pp. 1–721. [Google Scholar]
- Martin KA, Siefert JL, Yerrapragada S, Lu Y, McNeill TZ, Moreno PA, Weinstock GM, Widger WR, Fox GE. Cyanobacterial signatures genes. Photosynth Res. 2003;75:211–221. doi: 10.1023/A:1023990402346. [DOI] [PubMed] [Google Scholar]
- Lerat E, Daubin V, Moran NA. From Gene Trees to Organismal Phylogeny in Prokaryotes:The Case of the gamma-Proteobacteria. PLoS Biol. 2003;1:E19. doi: 10.1371/journal.pbio.0000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths E, Gupta RS. Distinctive protein signatures provide molecular markers and evidence for the monophyletic nature of the Deinococcus-Thermus phylum. J Bacteriol. 2004;186:3097–3107. doi: 10.1128/JB.186.10.3097-3107.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RS. The Phylogeny and Signature Sequences characteristics of Fibrobacters, Chlorobi and Bacteroidetes. Crit Rev Microbiol. 2004;30:123–143. doi: 10.1080/10408410490435133. [DOI] [PubMed] [Google Scholar]
- Gupta RS, Pereira M, Chandrasekera C, Johari V. Molecular signatures in protein sequences that are characteristic of Cyanobacteria and plastid homologues. Int J Syst Evol Microbiol. 2003;53:1833–1842. doi: 10.1099/ijs.0.02720-0. [DOI] [PubMed] [Google Scholar]
- Gupta RS, Griffiths E. Critical Issues in Bacterial Phylogenies. Theor Popul Biol. 2002;61:423–434. doi: 10.1006/tpbi.2002.1589. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein databases search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity GM, Holt JG. The road map to the manual. In: Boone DR and Castenholz RW, editor. Bergey's Manual of Systematic Bacteriology. 2nd. Berlin, Springer-Verlag; 2001. pp. 119–166. [Google Scholar]
- Dumler JS, Barbet AF, Bekker CP, Dasch GA, Palmer GH, Ray SC, Rikihisa Y, Rurangirwa FR. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and 'HGE agent' as subjective synonyms of Ehrlichia phagocytophila. Int J Syst Evol Microbiol. 2001;51:2145–2165. doi: 10.1099/00207713-51-6-2145. [DOI] [PubMed] [Google Scholar]
- Boussau B, Karlberg EO, Frank AC, Legault BA, Andersson SG. Computational inference of scenarios for alpha-proteobacterial genome evolution. Proc Natl Acad Sci U S A. 2004;101:9722–9727. doi: 10.1073/pnas.0400975101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepkowski T, Czaplinska M, Miedzinska K, Moulin L. The variable part of the dnaK gene as an alternative marker for phylogenetic studies of rhizobia and related alpha Proteobacteria. Syst Appl Microbiol. 2003;26:483–494. doi: 10.1078/072320203770865765. [DOI] [PubMed] [Google Scholar]
- Ludwig W, Klenk HP. Overview: A phylogenetic backbone and taxonomic framework for prokaryotic systamatics. In: Boone DR and Castenholz RW, editor. Bergey's Manual of Systematic Bacteriology. 2nd. Berlin, Springer-Verlag; 2001. pp. 49–65. [Google Scholar]
- Lerat E, Daubin V, Ochman H, Moran NA. Evolutionary Origins of Genomic Repertoires in Bacteria. PLoS Biol. 2005;3:e130. doi: 10.1371/journal.pbio.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulin L, Bena G, Boivin-Masson C, Stepkowski T. Phylogenetic analyses of symbiotic nodulation genes support vertical and lateral gene co-transfer within the Bradyrhizobium genus. Mol Phylogenet Evol. 2004;30:720–732. doi: 10.1016/S1055-7903(03)00255-0. [DOI] [PubMed] [Google Scholar]
- Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Mol Biol Evol. 2002;19:2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- Emelyanov VV. Common evolutionary origin of mitochondrial and rickettsial respiratory chains. Arch Biochem Biophys. 2003;420:130–141. doi: 10.1016/j.abb.2003.09.031. [DOI] [PubMed] [Google Scholar]
- Batut J, Andersson SG, O'Callaghan D. The evolution of chronic infection strategies in the alpha-proteobacteria. Nat Rev Microbiol. 2004;2:933–945. doi: 10.1038/nrmicro1044. [DOI] [PubMed] [Google Scholar]
- Daubin V, Ochman H. Bacterial genomes as new gene homes: the genealogy of ORFans in E. coli. Genome Res. 2004;14:1036–1042. doi: 10.1101/gr.2231904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang BF, Gray MW, Burger G. Mitochondrial genome evolution and the origin of eukaryotes. Annual Review of Genetics. 1999;33:351–397. doi: 10.1146/annurev.genet.33.1.351. [DOI] [PubMed] [Google Scholar]
- Emelyanov VV. Rickettsiaceae, rickettsia-like endosymbionts, and the origin of mitochondria. Biosci Rep. 2001;21:1–17. doi: 10.1023/A:1010409415723. [DOI] [PubMed] [Google Scholar]
- Kurland CG, Andersson SG. Origin and evolution of the mitochondrial proteome. Microbiol Mol Biol Rev. 2000;64:786–820. doi: 10.1128/MMBR.64.4.786-820.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser C, Ahmadinejad N, Wiegand C, Rotte C, Sebastiani F, Gelius-Dietrich G, Henze K, Kretschmann E, Richly E, Leister D, Bryant D, Steel MA, Lockhart PJ, Penny D, Martin W. A Genome Phylogeny for Mitochondria Among alpha-Proteobacteria and a Predominantly Eubacterial Ancestry of Yeast Nuclear Genes. Mol Biol Evol. 2004;21:1643–1660. doi: 10.1093/molbev/msh160. [DOI] [PubMed] [Google Scholar]
- Hrdy I, Hirt RP, Dolezal P, Bardonova L, Foster PG, Tachezy J, Embley TM. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature. 2004;432:618–622. doi: 10.1038/nature03149. [DOI] [PubMed] [Google Scholar]
- Dyall SD, Yan W, Delgadillo-Correa MG, Lunceford A, Loo JA, Clarke CF, Johnson PJ. Non-mitochondrial complex I proteins in a hydrogenosomal oxidoreductase complex. Nature. 2004;431:1103–1107. doi: 10.1038/nature02990. [DOI] [PubMed] [Google Scholar]
- Koski LB, Golding GB. The closest BLAST hit is often not the nearest neighbor. J Mol Evol. 2001;52:540–542. doi: 10.1007/s002390010184. [DOI] [PubMed] [Google Scholar]