Abstract
The Arabidopsis thaliana ASYMMETRIC LEAVES1 (AS1) and AS2 genes are important for repressing class I KNOTTED1-like homeobox (KNOX) genes and specifying leaf adaxial identity in leaf development. RNA-dependent RNA polymerases (RdRPs) are critical for posttranscriptional and transcriptional gene silencing in eukaryotes; however, very little is known about their functions in plant development. Here, we show that the Arabidopsis RDR6 gene (also called SDE1 and SGS2) that encodes a putative RdRP, together with AS1 and AS2, regulates leaf development. rdr6 single mutant plants displayed only minor phenotypes, whereas rdr6 as1 and rdr6 as2 double mutants showed dramatically enhanced as1 and as2 phenotypes, with severe defects in the leaf adaxial-abaxial polarity and vascular development. In addition, the double mutant plants produced more lobed leaves than the as1 and as2 single mutants and showed leaf-like structures associated on a proportion of leaf blades. The abnormal leaf morphology of the double mutants was accompanied by an extended ectopic expression of a class I KNOX gene BREVIPEDICELLUS (BP) and high levels of microRNA165/166 that may lead to mRNA degradation of genes in the class III HD-ZIP family. Taken together, our data suggest that the Arabidopsis RDR6-associated epigenetic pathway and the AS1-AS2 pathway synergistically repress BP and MIR165/166 for proper plant development.
INTRODUCTION
In higher plants, leaf primordia initiate from flanks of the shoot apical meristem (SAM) (Bowman et al., 2002). The initiation of leaf primordia coincides with the downregulation of the expression of several class I KNOTTED1-like homeobox (KNOX) genes (Byrne et al., 2000; Ori et al., 2000; Semiarti et al., 2001), which are required for the maintenance and growth of the SAM (Long et al., 1996; Bowman and Eshed, 2000; Volbrecht et al., 2000). Leaf development also requires the establishment of proximodistal, mediolateral, and adaxial-abaxial polarities (Hudson, 2000). The adaxial-abaxial polarity refers to the two opposing faces of a leaf blade, which have distinct cell types that have different biological functions (McConnell et al., 2001). It was reported that leaf primordia isolated from the SAM at the anlage stage could only form a small radial leaf without adaxial differentiation (Sussex, 1954, 1955). This result indicates that the establishment of the adaxial-abaxial axis is critical for subsequent asymmetric leaf development (Bowman et al., 2002).
In Arabidopsis thaliana, several genes have been identified for the formation of the adaxial-abaxial polarity, including class III HD-ZIP genes for promoting adaxial fate (McConnell and Barton, 1998; McConnell et al., 2001; Emery et al., 2003) and members of the YABBY and KANADI families for specifying leaf abaxial identity (Chen et al., 1999; Sawa et al., 1999; Siegfried et al., 1999; Eshed et al., 2001; Kerstetter et al., 2001). In addition, the ASYMMETRIC LEAVES1 (AS1) and AS2 genes are also responsible for establishing leaf polarity by specifying leaf adaxial identity (Xu et al., 2002, 2003). AS1 encodes a protein that contains an R2-R3 MYB domain (Byrne et al., 2000; Sun et al., 2002), suggesting that it may bind to DNA. AS2 encodes a plant-specific protein that can associate with AS1 (Iwakawa et al., 2002; Xu et al., 2002, 2003). These two genes have demonstrated roles in repressing class I KNOX genes (Byrne et al., 2000; Ori et al., 2000; Semiarti et al., 2001) and promoting leaf vein development (Semiarti et al., 2001; Sun et al., 2002). Recent studies have also revealed that in early leaf development, AS1 and AS2 are required for a proper auxin distribution and response (Zgurski et al., 2005). Additionally, AS1 and AS2 positively regulate the PHABULOSA (PHB) gene, a class III HD-ZIP family member (Lin et al., 2003; Xu et al., 2003). Although AS1 and AS2 functions have been studied extensively, it is unclear whether other genes function together with AS1 and AS2 for leaf patterning. If these genes exist, it would therefore be important to determine what roles they play in leaf development.
In both plants and animals, double-stranded RNA (dsRNA) that is processed to short RNAs (∼21 to 24 nucleotides) can trigger two coupled types of RNA-mediated gene silencing events: posttranscriptional gene silencing (PTGS) and transcriptional gene silencing (TGS) (Cogoni and Macino, 1999; Lipardi et al., 2001; Matzke et al., 2001; Nishikura, 2001; Sijen et al., 2001; Ahlquist, 2002; Aufsatz et al., 2002). Members of the RNA-dependent RNA polymerase (RdRP) family use small RNA molecules to prime dsRNA synthesis (Lipardi et al., 2001; Sijen et al., 2001) and have been defined as one of the important factors in PTGS and TGS. In recent years, genes encoding putative RdRPs have been isolated and characterized from several species, including tomato (Lycopersicon esculentum), Neurospora crassa, Caenorhabditis elegans, and Arabidopsis (Schiebel et al., 1993, 1998; Cogoni and Macino, 1999; Dalmay et al., 2000; Mourrain et al., 2000; Smardon et al., 2000). In Arabidopsis, mutations in the RDR6 gene, which encodes a putative RdRP protein, causes defects in PTGS (Dalmay et al., 2000; Mourrain et al., 2000). In addition, a more recent study revealed that the RDR6 gene is required for vegetative phase change (Peragine et al., 2004). However, because rdr6 mutant plants do not show severe morphology defects, the role of RdRPs in leaf pattern specification is largely unknown.
MicroRNAs (miRNAs) are endogenous ∼20- to 22-nucleotide RNAs that contribute important functions in animal and plant development by causing mRNA cleavage or protein translational repression (Bartel, 2004). The Arabidopsis miRNAs miR165 and miR166 differ by only one nucleotide and were found to share perfect complementarity with the regions of class III HD-ZIP genes encoding the START domain (Rhoades et al., 2002). A single nucleotide change within the START-encoding regions of some of these genes resulted in dominant mutations that caused aberrant adaxial-abaxial polarity and abnormal venation in leaves (McConnell et al., 2001; Emery et al., 2003; Zhong and Ye, 2004). Recent in vitro and in vivo experiments have shown that miR165/166 can indeed cause degradation of mRNAs of several HD-ZIP genes (Emery et al., 2003; Tang et al., 2003; Mallory et al., 2004; Zhong and Ye, 2004). Studies investigating the mechanisms of miRNA actions raise an important question: what gene(s) or pathway(s) regulates the level of miRNA gene expression in developmental processes? It was reported that the Arabidopsis gene ARGONAUTE1 (AGO1) both acts in and is regulated by the miRNA pathway (Kidner and Martienssen, 2004; Vaucheret et al., 2004). However, it is not clear whether other factors are also involved in the regulation of miRNA expression.
In this study, we describe the characterization of rdr6 single mutants and rdr6 as2 double mutants, which suggest a novel function of the previously reported RDR6 gene in leaf development. We report that the ectopic expression of a class I KNOX gene BREVIPEDICELLUS (BP) was extended and that miR165/166 levels were dramatically increased in the rdr6 as2 mutant leaves. We propose that the AS1-AS2 pathway and the RDR6-associated epigenetic pathway are both required for repression of BP and MIR165/166 in normal leaf development.
RESULTS
RDR6 Is Involved in Leaf Development
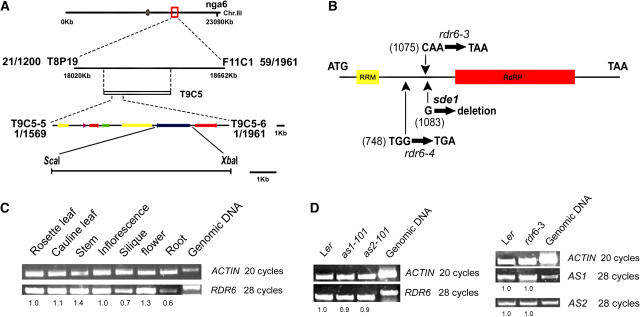
To identify additional genes functioning in the AS1-AS2 regulatory network, we conducted a genetic screen for enhancers and suppressors of the as2-101 mutant and identified two mutants showing similar enhanced as2 phenotypes. Genetic experiments revealed that these two mutants were allelic, and the single as2 enhancer mutant had nearly normal phenotypes (see Methods). We mapped this enhancer mutation to a 21-kb region on chromosome 3 (BAC T9C5), which contains seven predicted genes (Figure 1A). Sequencing of these candidate genes revealed that both of the as2 enhancer alleles carried mutations that introduce stop codons into the coding sequence of the RDR6, SDE1, or SGS2 genes (in this study, we refer to this gene as RDR6 for convenience), which encodes a previously reported RdRP protein (Figure 1B). Complementary experiments confirmed that mutations in the RDR6 gene caused an enhancement of as2 phenotypes (see Methods); we therefore renamed the as2 enhancer alleles as rdr6-3 as2-101 and rdr6-4 as2-101, respectively.
Figure 1.
Molecular Identification of the AS2 ENHANCER1 Gene.
(A) Fine-structure mapping. Seven putative genes (denoted by arrows) were identified between markers T9C5-5 and T9C5-6. The ScaI-XbaI genomic fragment containing the RDR6 gene was used in the complementation experiment.
(B) The RDR6 gene structure and positions of the nucleotide changes in rdr6-3, rdr6-4, and sde1-1 mutants. The yellow box indicates a region encoding the RNA recognition motif, and the red box represents a region encoding the RdRP domain (http://www.ncbi.nlm.nih.gov/BLAST).
(C) RT-PCR shows that RDR6 was expressed in all plant tissues examined.
(D) RDR6 expression was not affected by as1 and as2 mutations, and rdr6 mutation did not alter expression of the AS1 or AS2 genes. The numbers in (C) and (D) indicate the relative abundance of gene transcripts, which were calculated using the band intensity of the first lane as 1.0. as1-101, as2-101, and rdr6-3 are in the Ler genetic background.
RDR6 expression was found in all plant tissues examined (Figure 1C). The molecular genetics relationship between RDR6 and AS1/AS2 suggests that these genes may influence each other's expression. Therefore, we analyzed RDR6 expression in the as1-101 and as2-101 backgrounds and AS1 and AS2 expression in the rdr6-3 background (Figure 1D). The RT-PCR data revealed no obvious changes between the wild type and mutants, suggesting that direct transcriptional regulations do not occur between the RDR6 and AS1 and AS2 genes. These results indicate that the RDR6 gene may act separately from the AS1/AS2 pathway in the regulation of plant development.
Phenotypes of rdr6 Single Mutants and rdr6 as2 Double Mutants
To further understand the role of RDR6 in leaf development, we analyzed leaf phenotypes of rdr6-3 as2-101 plants. Compared with wild-type (Figure 2A) and as2-101 (Figure 2B) plants, rdr6-3 as2-101 (Figure 2C) and rdr6-4 as2-101 (Figure 2D) displayed severe phenotypes, with anthocyanin in some early-appearing rosette leaves. In addition, rdr6-3 as1-101 and rdr6-3 as2-101 double mutants showed similar phenotypes (Figure 2E). Because AS1 and AS2 can form protein complexes and may regulate the same downstream targets, the similar phenotypes in rdr6-3 as1-101 and rdr6-3 as2-101 suggest that the RDR6 gene or its product interacts with the AS1-AS2 pathway in leaf development. Because rdr6-3 as2-101 and rdr6-4 as2-101 exhibited similar overall phenotypes, we focused our additional phenotypic characterizations mainly on the rdr6-3 and rdr6-3 as2-101 mutants.
Figure 2.
Phenotypes of Single and Double Mutants.
(A) to (D) Morphological observations of wild-type and mutant seedlings. Wild-type Ler (A), as2-101 (B), rdr6-3 as2-101 (C), and rdr6-4 as2-101 (D).
(E) Statures of wild-type, as1-101, as2-101, rdr6-3, rdr6-3 as1-101, and rdr6-3 as2-101 plants at the early flowering stage. Note that rdr6-3 as1-101 and rdr6-3 as2-101 plants display similar phenotypes.
(F) rdr6-3 plants display distorted young rosette leaves (arrowhead).
(G) and (H) rdr6-3 (Ler ecotype) plants show some minor phenotypic changes.
(G) An additional inflorescence appears at the proximal end of some secondary inflorescences (arrowhead).
(H) Some cauline leaves are replaced by a filamentous structure (arrowhead).
(I) and (J) Phenotypic alterations of the previously reported sde1-1 mutant (Columbia-24 [Col-24] ecotype).
(I) An additional small inflorescence (arrowhead) that is similar to that of rdr6-3 in (G).
(J) A filamentous structure (arrowhead) appeared at the position where a cauline leaf should grow in the wild-type plants.
(K) A diagram summarizing the phenotypic changes observed in the rdr6-3 mutant. The rdr6-3 mutant shows a slightly reduced plant stature. Arrowheads indicate the following abnormalities: (1) additional inflorescence, (2) without cauline leaves or with a small filamentous structure, and (3) with some class IV inflorescences that were not seen in wild-type plants. Red circles indicate inflorescences.
All mutant plants except sde1 (Dalmay et al., 2000) in (I) and (J) are in the Ler genetic background. Bars = 5 mm in (A) to (J).
The rdr6-3 plants displayed only minor phenotypes, including a slightly reduced plant stature with slightly distorted young rosette leaves (Figure 2F) and an increased number of secondary inflorescences (8.9 ± 1.0 in the rdr6-3 mutant versus 5.9 ± 1.0 in the wild type, n = 20, P < 0.01). In addition, some rdr6-3 plants produced an extra small inflorescence at the base of some secondary inflorescences (Figure 2G; 34 small inflorescences/120 rdr6-3 plants; none were found in 100 wild-type plants). Furthermore, many rdr6-3 plants produced secondary inflorescences without a cauline leaf or with a variously sized filamentous structure at their proximal ends (Figure 2H; 118/242 rdr6-3 plants; 3/100 wild-type plants).
Analysis of the previously isolated sde1-1 single mutant (allelic to rdr6) revealed that although sde1-1 and rdr6-3 are from different ecotypes, they produce similar morphological abnormalities (Figures 2I and 2J; for comparison, see Figures 2G and 2H). In addition, plants with phenotypes similar to those of rdr6-3 as2-101 and rdr6-4 as2-101 mutant plants (Figures 2C and 2D) were observed in the F2 progeny of a cross between sde1-1 and as2-101 mutants (data not shown). Phenotypic analyses of the rdr6 mutant plants indicate that although rdr6 mutant phenotypes were relatively mild, the RDR6 gene has multiple functions in development (Figure 2K).
rdr6 as2 Exhibits Extended Ectopic Expression of BP
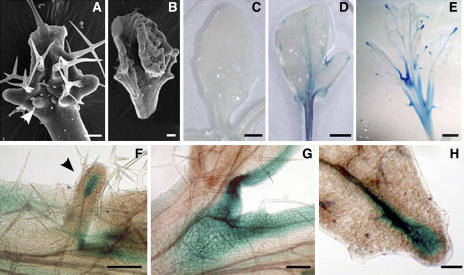
AS1 and AS2 repress the KNOX genes during leaf development, and leaves of some as1 and as2 alleles are serrated or even lobed because of the ectopic expressions of KNOX genes (Ori et al., 2000; Semiarti et al., 2001). Because the rdr6-3 as2-101 plants showed more lobed leaves (0 in 44 as2-101 plants and 32 in 44 rdr6-3 as2-101 plants), we examined by analyzing leaf morphology and expression of a class I KNOX gene BP in as2 and rdr6 as2 leaves whether RDR6 is also involved in repressing KNOX genes. We found that ectopic leaf primordia appeared frequently on the adaxial side of rdr6-3 as2-101 leaves (Figure 3A), and some of the ectopic leaf primordia eventually formed leaf-like structures (Figure 3B; 4 in 44 rdr6-3 as2-101 plants). To examine whether the expression of BP was altered in the rdr6-3 as2-101 leaves, we introduced a BP:β-glucuronidase (GUS) fusion into rdr6-3, as2-101, or rdr6-3 as2-101 plants by crosses and examined GUS staining in these mutant plants. BP was repressed in wild-type (Figure 3C) and rdr6-3 leaves (data not shown), whereas it was expressed in both as2-101 (Figure 3D) and rdr6-3 as2-101 leaves (Figure 3E), especially in sinuses of leaf lobes and leaf veins (Figures 3D, 3E, 3G, and 3H). In rdr6-3 as2-101 leaves, GUS staining was also observed in the ectopic leaf primordium (Figure 3F). Interestingly, BP was expressed strongly at the distal ends of each major leaf vein (Figures 3E and 3H) of the double mutants, but this pattern was not observed in the as2-101 single mutant (see Supplemental Figure 1 online). These BP expression observations suggest that RDR6 may negatively regulate BP in as2-101 mutant leaf blades.
Figure 3.
Phenotypes of Early Leaves and BP Expression in rdr6-3 as2-101 Mutant Plants.
(A) A young rdr6-3 as2-101 leaf showing primordium-like structures on the adaxial side (arrowhead).
(B) Some of the ectopic primordia on the leaf blade can eventually form a leaflet structure.
(C) Wild-type leaves carrying BP:GUS did not show any GUS activity.
(D) and (E) GUS staining was visible in sinuses of leaf lobes and major leaf veins in as2-101 (D) and rdr6-3 as2-101 mutants (E). Note that some rosette leaves of as2-101 in a mixed Col-Ler background contained lobes.
(F) to (H) Close-ups of GUS staining in rdr6-3 as2-101 mutant leaves.
(F) GUS activity was shown at the tip of a major vein (arrowhead) in an ectopic leaf primordium.
(G) GUS staining in sinuses of leaf lobes.
(H) GUS staining at the tip of a major leaf vein.
Leaves in (A) and (B) were from mutant plants in the Ler background, and leaves or leaf tissue in all other images were from the F2 plants of a cross between as2-101 rdr6-3 (Ler) and a transgenic line carrying the BP:GUS fusion (Col). Bars = 80 μm in (B), 100 μm in (F) and (H), 200 μm in (A) and (G), 2 mm in (C) and (D), and 1 mm in (E).
rdr6 as2 Has Severe Defects in Leaf Vein Formation and Adaxial-Abaxial Polarity
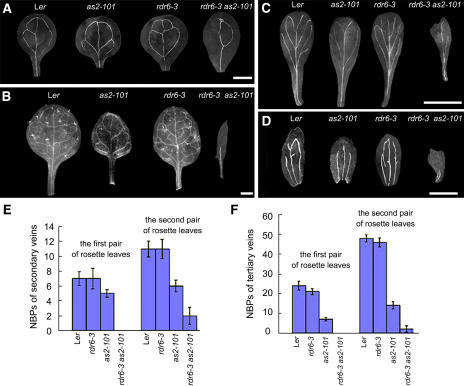
Leaf vein development is severely affected in the as1 and as2 mutants (Semiarti et al., 2001; Sun et al., 2002). To determine whether RDR6 is involved in regulating vein formation, we examined the leaves, sepals, and petals of the as2-101 and rdr6-3 as2-101 plants. Venations in all rdr6-3 leaves, sepals, and petals were similar to those in wild-type plants but showed a simple pattern in the as2-101 mutant (Figures 4A to 4D). The venation pattern in the rdr6-3 as2-101 mutant was even simpler than that of the as2-101 single mutant (Figures 4A to 4D). To gain more detailed information of leaf vein development, we analyzed the secondary and tertiary veins by scoring the number of branching points (NBPs) (Hamada et al., 2000). NBPs were found to be dramatically reduced in rdr6-3 as2-101 leaves, especially in the fine veins (Figures 4E and 4F). These results indicate that RDR6 is also involved in the vascular development of lateral organs.
Figure 4.
Abnormalities of Leaf Vascular Patterns of rdr6-3 as2-101 Mutant Plants.
(A) Venations of cotyledons in wild-type, as2-101, rdr6-3, and rdr6-3 as2-101 plants.
(B) Venations of the first rosette leaves in wild-type, as2-101, rdr6-3, and rdr6-3 as2-101 plants.
(C) Venations of petals in wild-type, as2-101, rdr6-3, and rdr6-3 as2-101 flowers.
(D) Venations of sepals in wild-type, as2-101, rdr6-3, and rdr6-3 as2-101 flowers. Note that venations of all leaves, sepals, and petals became very simple in the rdr6-3 as2-101 mutant plants.
(E) and (F) Quantitative analyses of NBPs in wild-type and rdr6-3 as2-101 leaves. Bars show standard deviation. Secondary leaf veins (E); tertiary leaf veins (F).
All mutants are in the Ler genetic background. Bars = 1 mm in (A) and (B) and 5 mm in (C) and (D).
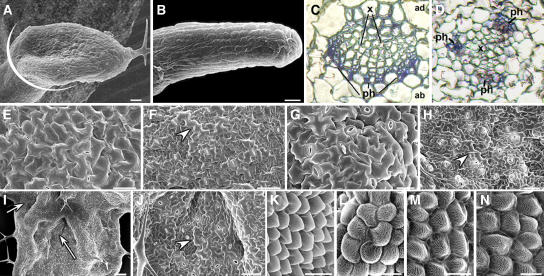
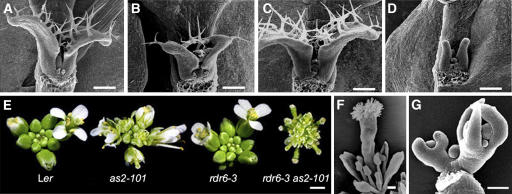
We reported previously that the as2-101 mutant is defective in leaf adaxial-abaxial polarity, showing abaxialized lotus- and needle-like leaves (Xu et al., 2002, 2003). To determine whether RDR6 plays a role in the adaxial-abaxial polarity formation of lateral organs, we analyzed leaf phenotypes of rdr6-3 as2-101 by scanning electron microscopy. Similar to the as2-101 mutant, we observed lotus-like (Figure 5A) and needle-like (Figure 5B) structures among rdr6-3 as2-101 rosette leaves but with a much higher frequency in rdr6-3 as2-101 than in as2-101 (90% versus 15% of the first-pair rosette leaves). The needle-like leaves in rdr6-3 as2-101 showed long and narrow epidermal cells (Figure 5B) similar to those of as2 (Xu et al., 2003) and phantastica (phan) (Waites and Hudson, 1995) but different from those of phb-1d (McConnell and Barton, 1998) and 35S:AS2 transgenic plants (Xu et al., 2003), suggesting that these leaves are abaxialized. In the as2-101 single mutants, lotus- and needle-like leaves only appeared in the first pair of rosette leaves. Strikingly, the rdr6-3 as2-101 plants showed lotus- and needle-like structures at all positions of rosette leaves, and even some cauline leaves were needle like (data not shown).
Figure 5.
Aberrant Adaxial-Abaxial Polarity in Leaves and Petals of the rdr6-3 as2-101 Mutant.
(A) A lotus leaf structure in rdr6-3 as2-101 plants. White line indicates the approximate position where sections were prepared for images in (C) and (D).
(B) A needle-like leaf in rdr6-3 as2-101 plants. Note that lotus- and needle-like leaves reflect a severe loss of adaxial-abaxial leaf polarity.
(C) and (D) Transverse sections through petioles near the blade.
(C) Wild-type Ler.
(D) rdr6-3 as2-101 double mutant. x, xylem; ph, phloem.
(E) to (J) Epidermal patterns of leaves.
(E) Adaxial epidermis of wild-type Ler.
(F) Abaxial epidermis of Ler.
(G) rdr6-3 as2-101 adaxial epidermis.
(H) rdr6-3 as2-101 abaxial epidermis.
(I) Patches appeared on the adaxial side of an rdr6-3 as2-101 leaf (arrows).
(J) A close-up of epidermal cells in a patch on the adaxial side of an rdr6-3 as2-101 leaf.
(K) to (N) Epidermal patterns of petals.
(K) Adaxial epidermis of Ler.
(L) Abaxial epidermis of Ler.
(M) Adaxial epidermis of rdr6-3 as2-101.
(N) Abaxial epidermis of rdr6-3 as2-101.
Arrowheads in (F), (H), and (J) point a long and irregularly shaped cell that reflects the abaxial characters of a leaf. All mutant plants are in the Ler genetic background. Bars = 25 μm in (A) and (B), 50 μm in (E), (F), (H), and (J), 100 μm in (I), 20 μm in (G), (K), and (M), and 10 μm in (L) and (N).
The vascular patterns in a series of sections of petioles at the position close to the blade were examined (Figure 5A, white line). In wild-type petioles, xylem develops on the adaxial pole of the vascular bundle, whereas phloem develops on the abaxial bundle pole (Figure 5C). By contrast, rdr6-3 as2-101 showed ectopic development of abaxial phloem tissue surrounding the xylem in petiole (Figure 5D), which is consistent with the view that rdr6-3 as2-101 leaves are abaxialized.
To further test the hypothesis that RDR6 function is required for adaxial-abaxial polarity formation of lateral organs, we analyzed leaf epidermal cells from the rdr6-3 as2-101 mutant. The adaxial epidermis of wild-type Landsberg erecta (Ler) leaves was characterized by an undulating surface composed of uniformly sized cells (Figure 5E), and the abaxial epidermis was characterized by smaller and less uniformly sized cells (Figure 5F; McConnell and Barton, 1998; Xu et al., 2003). The expanded leaves of as1 and as2 and all rdr6-3 leaves showed a similar epidermal pattern to that of wild-type plants (data not shown). By contrast, the adaxial side of rdr6-3 as2-101 mutant leaves contained both adaxial epidermal cells (Figure 5G) and patches of abaxialized cells (Figures 5I and 5J). Cells in these patches were similar to those on the abaxial side of both the wild-type (Figure 5F) and rdr6-3 as2-101 leaves (Figure 5H). The mosaic adaxial-abaxial cells on the adaxial side of a lamina were also observed in the phan mutant leaves in Antirrhinum majus (Waites and Hudson, 1995) but not seen in as1 or as2 alleles. Interestingly, petals of rdr6-3 as2-101 also demonstrated a severe defect of adaxial-abaxial polarity, which was not observed in the as2-101 and rdr6-3 single mutants (data not shown). Wild-type adaxial petal epidermis shows cone-shaped cells with straight lines (Figure 5K), whereas the abaxial petal epidermis shows cobblestone-shaped cells with wavy lines (Figure 5M). Adaxial epidermal cells in the rdr6-3 as2-101 petals (Figure 5L) were different from those in the wild type (Figure 5K) but rather similar to the abaxial epidermal cells of petals in both wild-type (Figure 5M) and rdr6-3 as2-101 double mutant plants (Figure 5N). Phenotypes of leaves and petals in the rdr6-3 as2-101 mutant plants further support the hypothesis that RDR6 function is required, along with the AS1-AS2 pathway, for adaxial-abaxial polarity formation.
In addition to the enhancement of as2 abnormalities observed in leaves and petals, the rdr6-3 as2-101 mutant also showed other abnormal phenotypes. These included a markedly slowed growth of leaf primordia (Figures 6A to 6D) and defective floral organs. The carpel surfaces were wrinkled, and sepals, petals, and stamens were small in size (Figures 6E and 6F). The aberrant floral organs formed at very early flower developmental stages, when narrowed sepals failed to enwrap the inner-whorl organs (Figure 6G). Mature rdr6-3 as2-101 flowers also showed some needle-like sepals and petals (data not shown). The pleiotropic rdr6-3 as2-101 phenotypes indicate that the RDR6 gene is required for both vegetative and reproductive developmental processes.
Figure 6.
rdr6-3 as2-101 Mutation Affects Other Developmental Processes.
(A) to (D) Six-day-old seedlings showed retarded rosette leaf growth in the rdr6-3 as2-101 mutant. Wild-type Ler (A); as2-101 (B); rdr6-3 (C); rdr6-3 as2-101 (D).
(E) Inflorescence phenotypes of wild-type, as2-101, rdr6-3, and rdr6-3 as2-101.
(F) An rdr6-3 as2-101 flower showing the short outer three whorls of organs.
(G) Abnormal flower phenotypes in the rdr6-3 as2-101 mutant appeared during very early developmental stages, when sepals failed to enwrap the inner floral organs.
All mutant plants are in the Ler genetic background. Bars = 80 μm in (A) to (D), (F), and (G) and 1 mm in (E).
miR165/166 Are Repressed by RDR6, AS1, and AS2
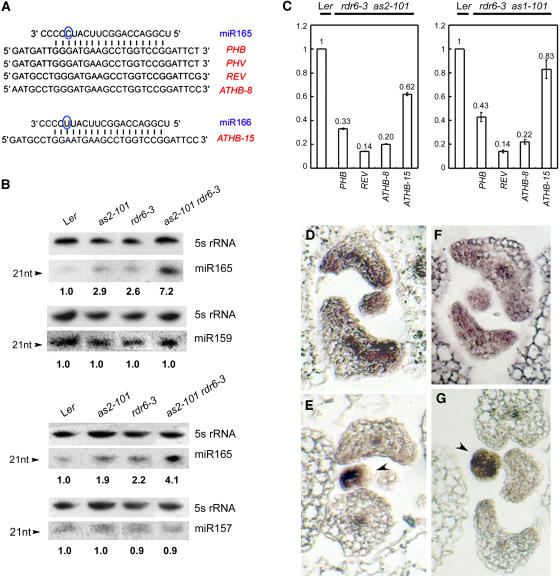
Both the differentiation of vascular tissue and the establishment of adaxial-abaxial polarity in Arabidopsis leaves are known to require the five class III HD-ZIP family genes: PHB, PHAVOLUTA (PHV), REVOLUTA (REV), ATHB8, and ATHB15 (Baima et al., 1995, 2001; McConnell and Barton, 1998; McConnell et al., 2001; Emery et al., 2003; Ohashi-Ito and Fukuda, 2003; Dinneny and Yanofsky, 2004). Four of these genes share a region that is complementary to the sequence of miR165, whereas the same region of ATHB15 matches the sequence of another miRNA, miR166 (Figure 7A). Because PHB is regulated by AS1 and AS2 (Lin et al., 2003; Xu et al., 2003) and RDR6 and AS1/AS2 synergistically regulate leaf adaxial-abaxial polarity and vascular development, we examined levels of miRNA165/166 and the five HD-ZIP transcripts in wild-type and rdr6-3 as2-101 mutant leaves.
Figure 7.
Analyses of Gene Expression.
(A) Sequences of miR165, miR166, and the region encoding the START domain of class III HD-ZIP proteins. Note that the circled C and U residues denote the nucleotide at which miR165 and miR166 differ.
(B) miRNA filter hybridizations. Lengths of RNA fragments in nucleotides (nt) and probes used in hybridizations are indicated on the left and right of each hybridization image, respectively. RNA gel blots were first probed by miRNAs, and the same filters were then analyzed by 5S RNA probe. Results were consistent from two separate experiments, using different preparations of RNA samples. Hybridization intensity was measured by phosphor image analysis, and the numbers indicate the relative abundance of miR165 levels, calculated using the band intensity of the first lane (Ler) as 1.0.
(C) Steady state levels of mRNA of class III HD-ZIP genes in rdr6-3 as1-101 (right) and rdr6-3 as2-101 (left). RNA was quantified for the indicated mRNA by real-time RT-PCR using primers surrounding the cleavage site. Quantification was normalized to that of ACTIN and then to the value of the wild-type plants, whose value was arbitrarily fixed at 1. Bars show standard error.
(D) and (E) In situ hybridization using an antisense PHB probe on transverse sections of leaf primordia in wild-type Ler (D) and rdr6-3 as2-101 (E).
(F) and (G) In situ hybridization using an antisense REV probe on transverse sections of leaf primordia in wild-type Ler (F) and rdr6-3 as2-101 (G). Arrowheads indicate the earlier-stage leaf primordia. All mutants analyzed are in the Ler genetic background.
Because miR165 and miR166 differ by only one nucleotide (Figure 7A) and molecular hybridization may not be able to distinguish one from the other, the hybridization signals with the miR165 probe probably reflect the levels of both transcripts. miR165/166 transcripts were detected at low levels in the wild-type Ler leaves, and their levels were increased slightly in as1-101, as2-101, and rdr6-3 single mutant leaves (Figure 7B). Strikingly, the miR165/166 level in rdr6-3 as1-101 and rdr6-3 as2-101 double mutants was dramatically elevated (Figure 7B), indicating that RDR6, AS1, and AS2 likely downregulate miR165/166. To test whether the RDR6, AS1, and AS2 regulation was specific for miR165/166, we examined levels of miR157 and miR159, which are also expressed in leaves (Reinhart et al., 2002). We found that the accumulation of these two miRNAs did not differ between the wild-type and mutant plants (Figure 7B), indicating that not all miRNAs are affected by RDR6, AS1, and AS2.
To determine whether increased miR165/166 levels can cause cleavage of transcripts of class III HD-ZIP genes in leaves, we performed RT-PCR using gene-specific primers spanning the predicted cleavage site. Compared with the real-time RT-PCR products from Ler leaves, those from rdr6-3 as2-101 (Figure 7C, left) and rdr6-3 as1-101 (Figure 7C, right) had decrease levels of PHB, REV, ATHB8, and ATHB15 transcripts, whereas the ATHB15 transcript, which does not completely match the miR165 sequence, was affected to a lesser extent. The PHV transcripts were not detected under our experimental conditions. To investigate whether the altered expression levels of these genes were also accompanied by changes in expression patterns, we performed PHB and REV in situ hybridization analyses. In wild-type plants, PHB and REV transcripts accumulated in the adaxial domain of a leaf primordium (Figures 7D and 7F). However, although PHB (Figure 7E) and REV (Figure 7G) transcripts were detected in the earlier-stage leaf primordium (arrowheads), they were reduced markedly in leaf primordia at the stage when leaf polarity began to establish. These results suggest that the increased miR165/166 levels may result in transcript degradations of the HD-ZIP genes.
Localization of miR165/166
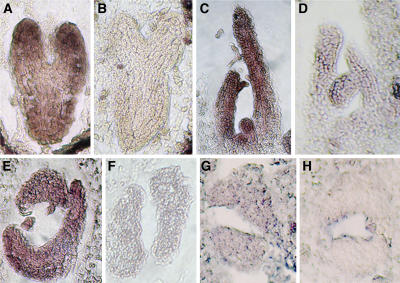
It was previously reported that miR165 was accumulated in the abaxial domain of leaves, so that only transcripts of class III HD-ZIP genes in the abaxial domain were eliminated, whereas those in the adaxial domain can normally function to promote the adaxial cell fates (Kidner and Martienssen, 2004). However, this model could not explain why the increased miR165/166 levels (supposedly in the abaxial domain) resulted in degradation of class III HD-ZIP transcripts that are located in the adaxial domain in rdr6-3 as2-101 leaves. To more clearly examine the localization of miR165/166 in leaf primordium, we performed in situ hybridizations using 4-concatenate copies of miR165 as a probe and following a protocol by Drews et al. (1991). At the torpedo stage, miR165/166 signals were detected throughout the entire embryo, with strongest hybridization in the distal parts of cotyledons and roots (Figure 8A). To our surprise, miR165/166 in the leaf primordium and young leaf did not show an abaxially accumulated pattern but was instead detected throughout the entire organ (Figure 8C). To ensure that this miR165/166 distribution pattern was correct, we repeated the in situ hybridization experiment using a different experimental protocol (Long and Barton, 1998). Again, miR165/166 signals were detected throughout the leaf primordia (Figure 8E). To avoid a potential artificial effect of the probe consisting of the 4-concatenate miR165 copies, we also used a sequence containing the predicted pre-miR165a sequence (Reinhart et al., 2002) as a probe to perform in situ hybridization experiments. Although the hybridization signals were very weak, they did not show a polar miR165/166 distribution (Figure 8G). miR165 sense probes did not produce hybridization signals under corresponding conditions (Figures 8B, 8D, 8F, and 8H). Taken together, our results reveal a nonpolar miR165/166 distribution pattern in leaf primordia, unlike that reported previously (Kidner and Martienssen, 2004).
Figure 8.
In Situ Localization of miR165/166 Expression in Wild-Type Ler Plants.
Hybridizations were performed with the antisense miR165 probe ([A], [C], [E], and [G]) or the sense miR165 probe ([B], [D], [F], and [H]). In (A) to (F), 4-concatenate miR165 was used as a probe. In (G) and (H), pre-miR165 sequence was used as a probe.
(A) and (B) Torpedo-stage embryos. miR165/166 were detected throughout the entire embryo in (A), but no signal was found in (B).
(C) and (D) Longitudinal sections of plant vegetative apices. miR165/166 were expressed in the SAM, leaf primordial, and young leaves in (C) but were undetectable in (D).
(E) and (F) Transverse sections of plant vegetative apices. miR165/166 hybridization signals were detected throughout the leaf primodia in (E), but no signal was detected in (F).
(G) Transverse sections of plant vegetative apices. miR165/166 signals were detected everywhere in the leaf primordium, though the signals were relatively weak.
(H) An antisense pre-165 could not detect miR165/166 signals.
FILAMENTOUS FLOWER Expression in rdr6-3 as2-101 Leaves
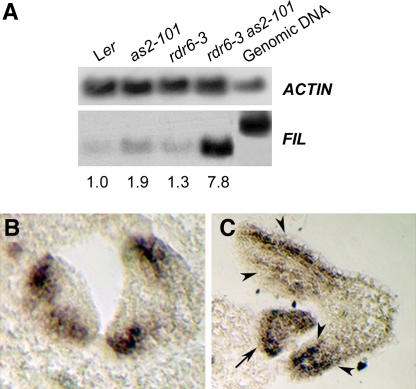
Our previous work has shown that rdr6-3 as2-101 mutant plants display abaxialized leaves (Figure 5) and the adaxial-promoting factors do not function properly (Figure 7). However, it is not known how abaxial-promoting factors, such as YABBY, might be affected in the double mutant leaves. To test for possible changes in YABBY gene functions in the abaxial fate of leaves, we analyzed the expression of a YABBY family member, FILAMENTOUS FLOWER (FIL), in the rdr6-3 as2-101 mutant by RT-PCR. In comparison with wild-type, as2-101, and rdr6-3 leaves, FIL transcripts were dramatically increased in the rdr6-3 as2-101 leaves (Figure 9A). FIL is usually expressed in the abaxial side of leaves (Figure 9B) but was extended throughout the entire primordium (Figure 9C, arrow) or to the adaxial side of young leaves (Figure 9C, arrowheads) in the rdr6-3 as2-101 double mutant. These results indicate that similar to BP and miR165/166, FIL is repressed by the AS1-AS2 and RDR6 pathways in leaf development.
Figure 9.
Expression of the FIL Gene.
(A) RT-PCR detection of FIL transcript levels in wild-type, as2-101, rdr6-3, and rdr6-3 as2-101 leaves. Numbers indicate the relative abundance of gene transcripts and were calculated using band intensity of the first lane (Ler) as 1.0.
(B) and (C) In situ hybridization using an antisense FIL probe to transverse sections of leaf primordia in wild-type Ler (B) and rdr6-3 as2-101 (C) that is in the Ler genetic background. Note that the FIL expression was extended to the entire leaf primordium (arrow) and the adaxial side of young leaves (arrowheads).
Overexpression of MIR165a
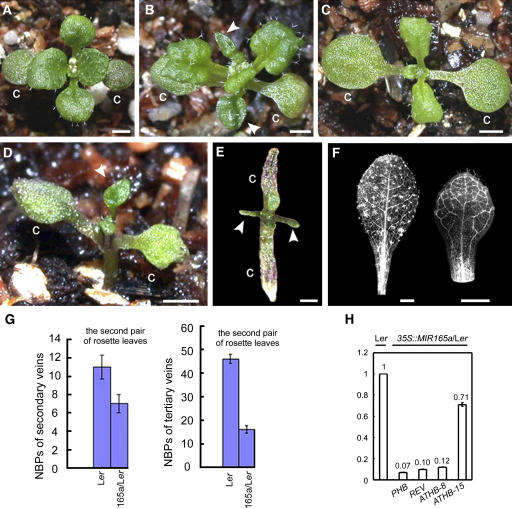
The Arabidopsis genome contains two genes for miR165: MIR165a and MIR165b (Reinhart et al., 2002). To test whether an elevated level of miR165 affects leaf polarity and vein formation in rdr6-3 as2-101, we introduced a 35S:MIR165a fusion into wild-type Ler, as2-101, and rdr6-3 plants. The increased miR165 levels in the transgenic plants were verified by RNA filter hybridization (see Supplemental Figure 2 online). The Ler background yielded 27 transgenic lines showing similar phenotypes of varying severity. Generally, the rosette leaves of these plants were smaller and round, with leaf margins slightly curled downward, resembling the margins of as1/as2 leaves (Figure 10A). The as2-101 background yielded 27 lines with similar phenotypes to each other (Figure 10B). The leaf shape was similar to that of as2-101 plants, but the leaf surfaces became ruffled, similar to those of rdr6-3 as2-101 double mutant leaves. In addition, these plants showed an increased frequency of lotus leaves compared with that in the as2-101 plants (62% versus 1% of the first-pair rosette leaves, in plants grown on plates at 22°C). In comparison, the first pair of rosette leaves in 22 35S:MIR165a/rdr6-3 transgenic lines resembled the expanded as2-101 leaves, with down-curled leaf margins (Figure 10C; for comparison, see Figure 2B). Two other 35S:MIR165a/rdr6-3 lines displayed even stronger leaf phenotypes, showing lotus leaf structures (Figure 10D) or needle-like leaves with anthocyanin accumulation (Figure 10E). In addition to the abnormal leaf polarity, transgenic lines also showed reduced leaf venation (Figures 10F and 10G).
Figure 10.
Phenotypes of 35S:MIR165a Transgenic Plants.
(A) A 35S:MIR165a/Ler seedling, showing small and round leaves with margins slightly curled downward. C, cotyledon.
(B) A 35S:MIR165a/as2-101 seedling with ruffled leaf surfaces. Arrowheads indicate the lotus leaves.
(C) A 35S:MIR165a/rdr6-3 seedling. First-pair rosette leaves resemble those of the as2-101 mutant plants.
(D) A 35S:MIR165a/rdr6-3 transgenic plant showing a lotus leaf structure (arrowhead).
(E) A 35S:MIR165a/rdr6-3 transgenic plant displaying severe phenotypes with abaxialized needle-like leaves (arrowheads).
(F) Comparison of venation between wild-type (left) and 35S:MIR165a/Ler (right) leaves, showing that venation of the 35S:MIR165a/Ler transgenic line was simplified. Bars = 1 mm in (A) to (F).
(G) Quantitative analyses of NBPs in wild-type and 35S:MIR165a/Ler transgenic plants.
(H) Real-time RT-PCR to examine steady state levels of mRNA of class III HD-ZIP genes in wild-type and 35S:MIR165a/Ler plants. Bars show standard error.
To investigate whether the altered leaf phenotypes in the transgenic plants are due to cleavage of class III HD-ZIP transcripts in a manner similar to that in the rdr6-3 as2-101 mutant, we performed real-time RT-PCR using rosette leaves of the T2 generation of 35S:MIR165a/Ler transgenic plants. In comparison with wild-type plants, the levels of PHB, REV, ATHB8, and ATHB15 transcripts in the 35S:MIR165a/Ler were reduced to varying degrees (Figure 10H), similar to those in rdr6-3 as2-101 and rdr6-3 as1-101 mutant plants (Figure 7C). The results from the 35S:MIR165a transgenic plants strongly support the idea that an increased level of miR165 can affect leaf polarity and venation.
DISCUSSION
RdRP is an enzyme that uses single-stranded RNAs as templates to synthesize dsRNAs (Lipardi et al., 2001; Matzke et al., 2001; Nishikura, 2001; Sijen et al., 2001). Cleavage of dsRNAs by RNase III helicase (Dicer) generates small interfering RNAs (siRNAs), which are recruited by the RNA-induced silencing complex for PTGS/TGS of their cognate genes (Vaucheret et al., 2001; Aufsatz et al., 2002; Hutvagner and Zamore, 2002; Vaistij et al., 2002; Volpe et al., 2002). The biological functions of RDR6 were previously thought to be associated with transgene silencing in transgenic plants (Dalmay et al., 2000; Mourrain et al., 2000; Vaistij et al., 2002) and virus resistance (Mourrain et al., 2000; Ahlquist, 2002). In this study, we demonstrate that the RDR6 function is also required for the specification of pattern in the leaf. The fact that RDR6 is involved in the mechanism for enhancing the as1/2 mutant phenotype suggests that PTGS/TGS may be required for normal plant development.
as1 and as2 mutant plants produce abaxialized leaves (Sun et al., 2002; Xu et al., 2002, 2003). We reported previously that expression of the PHB gene was enhanced in 35S:AS1/Ler and 35S:AS2/Ler transgenic plants and that expression of the FIL gene was elevated in as1-101 and as2-101 mutant plants (Xu et al., 2003). These results, together with other data (Lin et al., 2003; Engstrom et al., 2004), suggest that AS1 and AS2 are genetically upstream of the PHB and FIL genes in the regulation of leaf polarity. However, it was not clear how AS1 and AS2 might influence these downstream genes for leaf polarity formation. In this work, we propose that (1) AS1 and AS2 upregulate the PHB and REV genes for specifying leaf adaxial identity via the repression of miR165/166, and (2) the as1 and as2 phenotypes might be caused at least partially by the altered transcript levels of HD-ZIP III genes.
This hypothesis is supported by two lines of evidence. First, miR165/166 levels were elevated in the as1 and as2 single mutants and were dramatically increased in the rdr6 as1 and rdr6 as2 double mutants. These miR165/166 accumulations may be accompanied by cleavage of transcripts from class III HD-ZIP family members. Second, overexpression of MIR165a in the as2-101 and rdr6-3 single mutants resulted in a more severe defect in adaxial-abaxial leaf polarity than that seen in the as2-101 single mutant, indicating that the phenotype is a direct consequence of the change in miR165/166 content. Together, these data indicate that miRNA is an important link between AS1-AS2 and class III HD-ZIP genes in the regulatory network of leaf development.
The proposed model that miR165 is accumulated in the abaxial domain of leaf primordium (Kidner and Martienssen, 2004) cannot explain some observations about class III HD-ZIP expression levels. For example, the PHB transcript level in the phb-1d mutant was elevated throughout the leaf, indicating that miR165/166 activities exist in the adaxial domain of leaf primordia (McConnell et al., 2001; Bao et al., 2004). Furthermore, the expression patterns of each class III HD-ZIP gene are distinguishable, with the REV transcripts in the more expanded range in the adaxial side of lateral organs than those of PHB and PHV (Emery et al., 2003) and the ATHB-8 transcripts only in the vasculature (Baima et al., 1995). These results suggest that miR165/166 are not the sole factor affecting HD-ZIP distribution.
In this study, we report that miR165/166 are distributed throughout the leaf primordium. It is possible that miR165/166 expression may change over developmental stages of leaves. For example, according to the previous report, the miR165/166 distribution in P1-stage primordia seemed different from that in P2- and P3-stage primordia (Kidner and Martienssen, 2004). In fact, the miR165/166 distribution pattern in P1-stage primordia is similar to that of our in situ hybridization results, with signals in both adaxial and abaxial sides. In the older P2 and P3 primordia, signals became concentrated in the abaxial cells (Kidner and Martienssen, 2004). Additionally, we investigated whether the signal that is strongly accumulated in the cell walls of the abaxial domain of P2- and P3-stage primordia (Kidner and Martienssen, 2004) reflects a real cellular signal. Many previous in situ hybridization experiments using different probes indicated that signals at these stages of leaf primordia appear in the cytoplasm instead of in the cell walls (Bowman and Eshed, 2000; Byrne et al., 2000; Kerstetter et al., 2001; McConnell et al., 2001; Emery et al., 2003).
Our 35S:MIR165a transgenic results showed that in addition to its function in the leaf polarity formation, miR165 may have other regulatory functions during leaf development, including vascular development and leaf shape control. These data also imply that miR165 may be required for the development of the entire leaf, not only the abaxial leaf domain. The Arabidopsis genome contains two miR165 and seven miR166 copies (Rhoades et al., 2002). It will be important in the future to determine the expression pattern for each of these miRNAs by miRNA promoter:reporter fusions, which may improve our understanding of their roles in leaf development.
We observed dramatically enhanced FIL gene expression in rdr6-3 as2-101 mutant plants, with an expanded expression domain. Although it was previously proposed that the PHB pathway negatively regulates YABBY genes (Siegfried et al., 1999; Eshed et al., 2001; Bowman et al., 2002), our data suggest that the enhanced FIL expression may result from reduced HD-ZIP activity in the rdr6-3 as2-101 mutant plants. Alternatively, the increased FIL transcripts in the rdr6-3 as2-101 leaves may also result from defective PTGS/TGS, similar to those of BP and miR165/166.
Although it has been well documented that miRNAs play important roles in gene regulation in plants and animals, very little is known about how miRNA genes are regulated. Our data provide important information indicating that the AS1-AS2 pathway and the RDR6 pathway are both required for miR165/166 regulation. The RDR6 and AS1-AS2 pathways both negatively regulate BP and MIR165/166, but these regulations appeared to be processed separately. This idea was supported by the observation that RDR6 expression in the as1 and as2 mutants and AS1 and AS2 expression in the rdr6 mutant were not obviously different from that in wild-type plants. However, the rdr6 as2 double mutant showed much more severe phenotypes than either of the single mutants, indicating that the AS1-AS2 and RDR6 pathways may synergistically repress BP and MIR165/166 in leaves. It is not clear how these two pathways act to regulate their downstream targets. One possibility is that in addition to the downregulation of BP and MIR165/166 through the DNA binding function, AS1-AS2 may also be involved directly or indirectly in the generation of siRNAs used in the RDR6 pathway to repress BP and MIR165/166. Alternatively, these two proteins might interact with or affect the regulation of AGO1, which is known to play a role in both the miRNA and siRNA pathways. AS1-AS2 functions may be so critical that plants with a loss of function in the RDR6 gene do not show severe plant phenotypes. Although the AS1-AS2 pathway is disrupted in the as1 and as2 mutants, the RDR6 pathway can partially repress BP and MIR165/166. This regulation then attenuates the severity of plant phenotypes caused by as1 and as2 mutations. Plants with loss of function in both AS1-AS2 and RDR6 result in the abnormal BP and MIR165/166 actions, leading to leaves with more severe lobes, ectopic leaf primordia, and abnormal adaxial-abaxial polarity and venation.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana as1-101 and as2-101 mutants in the Ler background were generated as described previously (Sun et al., 2000, 2002; Xu et al., 2002). Seeds of sde1 (Col-24) and a BP:GUS transgenic line (Col) were kindly provided by D. Baulcombe (John Innes Centre, Norwich, UK) and S. Hake (University of California, Berkeley, CA), respectively. For generation of as2 enhancers, as2-101 seeds (Ler) were ethyl methanesulfonate mutagenized (0.1%), and ∼40,000 M2 plants from 1395 M1 lines were screened. To test whether the enhanced as2 phenotypes were due to a second site mutation, we pollinated the new mutant flowers with wild-type Ler pollen. The F1 progeny showed wild-type phenotypes, and F2 plants segregated with a distribution of 295 wild type, 78 as2-101, and 27 enhanced mutants, close to a 12:3:1 ratio. This suggested that the phenotypic enhancement was due to a mutation in an unlinked gene and that the new mutation alone did not cause obvious phenotypic changes. We then analyzed F3 plants of F2 wild-type-like plants and identified 16 F3 families that did not segregate for as2 mutants. We crossed one plant from each of these 16 families to as2-101. From these 16 crosses, one parent plant, whose 13 independent F2 families all yielded approximately one-sixteenth enhanced mutants, was considered to be the homozygous single as2-enhancing mutant. This plant was named as2 enhancer1 (ae1-1), and only this plant or its progeny was used for further work. The ae1-1 as2-101 mutant was backcrossed to wild-type Ler five times before phenotypic analysis.
Allelism tests between the two enhancer mutants were performed by pollinating the ae1-1 flowers with pollen from F1 plants of a cross between the second mutant and Ler. A total of 91 F1 plants consisted of 47 wild-type-like plants, 24 as2-101 mutants, and 20 enhanced plants, indicating that the two enhancer mutations were allelic. Therefore, the second as2 enhancer mutant was designated ae1-2 as2-101.
Map-Based Cloning
Mapping of the AE1 locus was performed by analysis of an F2 population from a cross between ae1-1 as2-101 and the polymorphic Col ecotype. The AE1 locus was mapped to the proximal arm of chromosome 3, near the simple sequence length polymorphic marker nga6. A set of simple sequence length polymorphic markers was used to detect polymorphisms between the Col and Ler ecotypes, and the AE1 locus was mapped to a 21-kb region (BAC T9C5), which contains seven predicted genes. Sequencing of these candidate genes from ae1-1 as2-101 and ae1-2 as2-101 revealed that a gene called RDR6 (also called SDE1 or SGS2) contained nucleotide substitutions that resulted in premature stop codons in both alleles.
For the complementation experiment, a 6.85-kb ScaI-XbaI genomic fragment containing the 4.0-kb RDR6 gene plus 0.85 and 2.0 kb of upstream and downstream sequences, respectively, was isolated from TAC clone K1J14 and inserted into the pCAMBIA1301 transformation vector. We then transformed ae1-1 mutant plants with this 6.85-kb RDR6 fragment and examined the transgenic T1 plants for complementation of the ae1 phenotype (the distorted young rosette leaves that appear in almost all ae1 plants). Eleven independent transgenic lines were obtained, and seven of those showed complete rescue of ae1 phenotypes. The transgenic plants were verified by PCR with one vector-specific primer (5′-TGATGGCATTTGTAGGAGC-3′) and one RDR6-specific primer (5′-TGCCCGAGAATCCAAATC-3′). Therefore, the ae1-1 as2-101 and ae1-2 as2-101 mutants were renamed rdr6-3 as2-101 and rdr6-4 as2-101, respectively.
RT-PCR
RNA extraction was performed as described previously (Xu et al., 2003) with leaves from 20-d-old seedlings, and reverse transcription was performed with 1 μg total RNA using a kit (Fermentas, Vilnius, Lithuania). PCR was performed in the presence of the double-stranded DNA-specific dye SYBR green (Shenyou, Shanghai, China) following the manufacturer's instructions. Amplification was monitored in real time with the fluorometric thermal cycler Rotor-Gene 2000 (Corbett Research, Sydney, Australia). PCR was performed with the following gene-specific primers: 5′-TGTGGAGAATGGAACCAC-3′ and 5′-CTAGCAGAGTTCCTTTCC-3′ for PHB (exon4/exon7), 5′-ATCTGTGGTCACAACTCC-3′ and 5′-TAGCGACCTCTCACAAAC-3′ for REV (exon3/exon8), 5′-GCTACCACAGATACTAGC-3′ and 5′-TCGCAAGGTCTAATGAGG-3′ for ATHB-8 (exon3/exon8), 5′-TCAAAGGCAACTGGAACC-3′ and 5′-GTGCAAGTACTTTGGGTG-3′ for ATHB-15 (exon4/exon9), and 5′-TGGCATCA(T/C)ACTTTCTACAA-3′ and 5′-CCACCACT(G/A/T)AGCACAATGTT-3′ for ACTIN. For real-time PCR, quantifications of each cDNA sample were made in triplicate, and the consistent results from at least two separately prepared RNA samples were used. For each quantification, conditions were, as recommended, 1 ≥ E ≥ 0.80 and r2 ≥ 0.980, where E is the PCR efficiency and r2 corresponds to the correlation coefficient obtained with the standard curve. For each quantification, a melt curve was realized at the end of the amplification experiment by steps of 0.5°C from 55 to 99°C. Results were normalized to that of ACTIN. PCR experiments for AS1, AS2, and RDR6 were performed with the following primers: 5′-CTGCGCCTCAACCGCCAATC-3′ and 5′-CCTTACATTACATTACAAGTTAC-3′ for AS1, 5′-TCCTCCGGCGAAAATGTC-3′ and 5′-CCGGCGAGTAAGTTGATGC-3′ for AS2, 5′-GGGACCTGTACTTTGTGGCTTGG-3′ and 5′-GGGCATGGACCAGATGTGACCC-3′ for RDR6 (exon1/exon2), and 5′-CTTACTTCAATCCCCAGG-3′ and 5′-CTTTTGGACATGATAAACCC-3′ for FIL (exon2/exon7). The PCR products were then analyzed by gel electrophoresis, and the relative abundance of gene transcripts was calculated by the GIS-2016 image system (Tanon, Shanghai, China) using the first-lane product as 1.0.
Construction of MIR165a Transgenic Plants
For overexpression of MIR165, a 141-bp MIR165a precursor was PCR amplified using genomic DNA from Ler plants with gene-specific primers 5′-GGGTTAAGCTATTTCAGTTG-3′ and 5′-AGAGGCAATAACATGTTGG-3′, confirmed by sequencing, and inserted into the pMON530 vector, downstream of the 35S promoter. This construct was introduced into Ler, as2-101, and rdr6-3 plants by Agrobacterium tumefaciens–mediated transformation. The transgenic lines were verified by PCR using a 35S-specific primer (5′-GCTCCTACAAATGCCATCA-3′) and a primer matching the MIR165a precursor sequence (5′-AGAGGCAATAACATGTTGG-3′). miR165 overexpression was confirmed by miRNA filter hybridization (see Supplemental Figure 2 online).
miRNA Filter Hybridization
Total RNA was extracted as described previously (Huang et al., 1995), and ∼5 to 10 μg RNA per lane was separated on a denaturing 19% polyacryamide gel (18 × 16 cm) containing 8 M urea, with each of the sense strands used as a positive control. Antisense probes (5′-GGGGGATGAAGCCTGGTCCGA-3′ for miR165, 5′-GTGCTCTCTATCTTCTGTCAA-3′ for miR157, and 5′-TAGAGCTCCCTTCAATCCAAA-3′ for miR159) were 32P-end labeled, and hybridization was performed according to the method described by Chen (2004).
Detection of GUS Activity and in Situ Hybridization
Histochemical detection of GUS activity was performed with 5-bromo-4-chloro-3-indolyl β-d-glucuronic acid (X-Gluc) as a substrate. Leaf tissue was placed in X-Gluc solution [750 μg/mL X-Gluc, 100 mM NaPO4, pH 7, 3 mM K3F3(CN)6, 10 mM EDTA, and 0.1% Nonidet P-40] under a vacuum for 10 min at room temperature, then incubated overnight at 37°C. In situ hybridizations were performed as previously described (Drews et al., 1991; Long and Barton, 1998) using 10-d-old seedlings. The probes were made from cDNA clones containing sequences from exons 4 to 7 for PHB, exons 2 to 7 for FIL, and exons 3 to 8 for REV or from a pGEM7Z(+) plasmid harboring a fragment of the 4-concatenate miR165 or a fragment of the predicted pre-miR165a (Reinhart et al., 2002).
Microscopy
Fresh tissue from wild-type and mutant plants was examined using a SZH10 dissecting microscope (Olympus, Tokyo, Japan) and photographed using a Nikon E995 digital camera (Nikon, Tokyo, Japan). Wild-type and mutant plant tissue was observed by scanning electron microscopy as previously described (Chen et al., 2000). Vascular patterns and vein numbers were analyzed with a Zeiss dissecting microscope (Jena, Germany) using the dark-field setting, according to our previously described methods (Sun et al., 2002).
Supplementary Material
Acknowledgments
The authors thank D. Baulcombe and Plant Bioscience Ltd. for the sde1-1 seeds, S. Hake for the BP:GUS transgenic seeds, Q. Lin and L. Pi for the rdr6-4 as2-101 seeds, The Ohio State University Arabidopsis Stock Center for TAC clone K1J14, X. Gao, J. Mao, H. Dai, and Y. Dou for their assistance with the scanning electron microscopy, and H. Ma, S. Luan, X. Chen, G. Tang, and W. Shen for helpful discussions and critical reading of the manuscript. This research was supported by grants from the Chinese Administration of Science and Technology (863), the Chinese National Scientific Foundation (30421001, 30370751, and 90208009), and the Shanghai Scientific Committee to H.H.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Hai Huang (hhuang@sippe.ac.cn).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.033449.
References
- Ahlquist, P. (2002). RNA-dependent RNA polymerase, viruses, and RNA silencing. Science 296 1270–1273. [DOI] [PubMed] [Google Scholar]
- Aufsatz, W., Mette, M.F., Winden, J.V.D., Matzke, A.J.M., and Matzke, M. (2002). RNA-directed DNA methylation in Arabidopsis. Proc. Natl. Acad. Sci. USA 99 16499–16506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baima, S., Nobili, F., Sessa, G., Lucchetti, S., Ruberti, I., and Morelli, G. (1995). The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121 4171–4182. [DOI] [PubMed] [Google Scholar]
- Baima, S., Possenti, M., Matteucci, A., Wisman, E., Altamura, M.M., Ruberti, I., and Morelli, G. (2001). The Arabidopsis ATHB-8 HD-Zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 126 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, N., Lye, K.-W., and Barton, M.K. (2004). MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev. Cell 7 653–662. [DOI] [PubMed] [Google Scholar]
- Bartel, D.V. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116 281–297. [DOI] [PubMed] [Google Scholar]
- Bowman, J., and Eshed, Y. (2000). Formation and maintenance of the shoot special meristem. Trends Plant Sci. 5 110–115. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Eshed, Y., and Baum, S.F. (2002). Establishment of polarity in angiosperm lateral organs. Trends Genet. 18 134–141. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Barley, R., Curtis, M., Arroyo, J.M., Dunham, M., Hudson, A., and Martienssen, R.A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408 967–971. [DOI] [PubMed] [Google Scholar]
- Chen, C., Wang, S., and Huang, H. (2000). LEUNIG has multiple functions in gynoecium development in Arabidopsis. Genesis 26 42–54. [DOI] [PubMed] [Google Scholar]
- Chen, Q., Atkinson, A., Otsuga, D., Christensen, T., Reynolds, L., and Drews, G.N. (1999). The Arabidopsis FILAMENTOUS FLOWER gene is required for flower formation. Development 126 2715–2726. [DOI] [PubMed] [Google Scholar]
- Chen, X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni, C., and Macino, G. (1999). Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399 166–169. [DOI] [PubMed] [Google Scholar]
- Dalmay, T., Hamilton, A., Rudd, S., Angell, S., and Baulcombe, D.C. (2000). An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101 543–553. [DOI] [PubMed] [Google Scholar]
- Dinneny, J.R., and Yanofsky, M.F. (2004). Vascular patterning: Xylem or phloem? Curr. Biol. 14 R112–R114. [PubMed] [Google Scholar]
- Drews, G.N., Bowman, J.L., and Meyerowtiz, E.M. (1991). Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65 991–1002. [DOI] [PubMed] [Google Scholar]
- Emery, J.F., Floyd, S.K., Alvarez, J., Eshed, Y., Hawker, N.P., Izhaki, A., Baum, S.F., and Bowman, J.L. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANIDI genes. Curr. Biol. 13 1768–1774. [DOI] [PubMed] [Google Scholar]
- Engstrom, E.M., Izhaki, A., and Bowman, J.L. (2004). Promoter bashing, microRNAs, and Knox genes. New insights, regulators, and targets-of-regulation in the establishment of lateral organ polarity in Arabidopsis. Plant Physiol. 135 685–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., Perea, L.V., and Bowman, J.L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11 1251–1260. [DOI] [PubMed] [Google Scholar]
- Hamada, S., Onouchi, H., Tanaka, H., Kudo, M., Liu, Y., Shibata, D., Machida, C., and Machida, Y. (2000). Mutations in the WUSCHEL gene of Arabidopsis thaliana result in the development of shoots without juvenile leaves. Plant J. 24 91–101. [DOI] [PubMed] [Google Scholar]
- Huang, H., Tudor, M., Weiss, C.A., Hu, Y., and Ma, H. (1995). The Arabidopsis MADS-box gene AGL3 is widely expressed and encodes a sequence-specific DNA-binding protein. Plant Mol. Biol. 28 549–567. [DOI] [PubMed] [Google Scholar]
- Hudson, A. (2000). Development of symmetry in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51 349–370. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G., and Zamore, P.D. (2002). A microRNA in a multiple-turnover RNAi enzyme complex. Science 297 2056–2060. [DOI] [PubMed] [Google Scholar]
- Iwakawa, H., Ueno, Y., Semiarti, E., Onouchi, H., Kojima, S., Tsukaya, H., Hasebe, M., Soma, T., Ikezaki, M., Machida, C., and Machida, Y. (2002). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43 467–478. [DOI] [PubMed] [Google Scholar]
- Kerstetter, R.A., Bollman, K., Taylor, R.A., Bomblies, K., and Poethlg, R.S. (2001). KANADI regulates organ polarity in Arabidopsis. Nature 411 706–709. [DOI] [PubMed] [Google Scholar]
- Kidner, C.A., and Martienssen, R.A. (2004). Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428 81–84. [DOI] [PubMed] [Google Scholar]
- Lin, W., Shuai, B., and Springer, P.S. (2003). The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 15 2241–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipardi, C., Wei, Q., and Paterson, B.M. (2001). RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell 107 297–307. [DOI] [PubMed] [Google Scholar]
- Long, J., and Barton, M.K. (1998). The development of apical embryonic pattern in Arabidopsis. Development 125 3027–3035. [DOI] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 66–69. [DOI] [PubMed] [Google Scholar]
- Mallory, A.C., Reinhart, B.J., Jones-Rhoades, M.W., Tang, G., Zamore, P.D., Barton, M.K., and Bartel, D.P. (2004). MicroRNA control of PHABULOSA in leaf development: Importance of pairing to the microRNA 5′ region. EMBO J. 23 3356–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke, M., Matzke, A.J.M., and Kooter, J.M. (2001). RNA: Guiding gene silencing. Science 293 1080–1083. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., and Barton, M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125 2935–2942. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411 709–713. [DOI] [PubMed] [Google Scholar]
- Mourrain, P., et al. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101 533–542. [DOI] [PubMed] [Google Scholar]
- Nishikura, K. (2001). A short primer on RNAi: RNA-directed RNA polymerase acts as a key catalyst. Cell 107 415–418. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito, K., and Fukuda, H. (2003). HD-Zip III homeobox genes that include a novel member, ZeHB-13 (Zinnia)/ATHB-15 (Arabidopsis), are involved in procambium and xylem cell differentiation. Plant Cell Physiol. 44 1350–1358. [DOI] [PubMed] [Google Scholar]
- Ori, N., Eshed, Y., Chuck, G., Bowman, J., and Hake, S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127 5523–5532. [DOI] [PubMed] [Google Scholar]
- Peragine, A., Yoshikawa, M., Wu, G., Heidi, L., and Poethig, R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart, B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. (2002). MicroRNAs in plants. Genes Dev. 16 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M.W., Reinhart, B.J., Lim, L.P., Burge, C.B., Bartel, B., and Bartel, D.P. (2002). Prediction of plant microRNA targets. Cell 110 513–520. [DOI] [PubMed] [Google Scholar]
- Sawa, S., Watanabe, K., Goto, K., Kanaya, E., Morita, E.H., and Okada, K. (1999). FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HNG-related domains. Genes Dev. 13 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiebel, W., Haas, B., Marinkovic, S., Klanner, A., and Sanger, H.L. (1993). RNA-directed RNA polymerase from tomato leaves. I. Purification and physical properties. J. Biol. Chem. 268 11851–11857. [PubMed] [Google Scholar]
- Schiebel, W., Pelissier, T., Riedel, L., Thalmeir, S., Schiebel, R., Kempe, D., Lottspeich, F., Sanger, H.L., and Wassenegger, M. (1998). Isolation of an RNA-directed RNA polymerase-specific cDNA clone from tomato. Plant Cell 10 2087–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semiarti, E., Ueno, Y., Tsukaya, H., Iwakawa, H., Machida, C., and Machida, Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128 1771–1783. [DOI] [PubMed] [Google Scholar]
- Siegfried, K.R., Eshed, Y., Baum, S.F., Otsuga, D., Drews, G.N., and Bowman, J. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126 4117–4128. [DOI] [PubMed] [Google Scholar]
- Sijen, T., Fleenor, J., Simmer, F., Thijssen, K.L., Parrish, S., Timmons, L., Plasterk, R.H.A., and Fire, A. (2001). On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107 465–476. [DOI] [PubMed] [Google Scholar]
- Smardon, A., Spoerke, J.M., Stacey, S.C., Klein, M.E., Mackin, N., and Maine, E.M. (2000). EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 10 169–178. [DOI] [PubMed] [Google Scholar]
- Sun, Y., Zhang, W., Li, F., Guo, Y., Liu, T., and Huang, H. (2000). Identification and genetic mapping of four novel genes that regulate leaf development in Arabidopsis. Cell Res. 10 325–335. [DOI] [PubMed] [Google Scholar]
- Sun, Y., Zhou, Q., Zhang, W., Fu, Y., and Huang, H. (2002). ASYMMETRIC LEAVES1, an Arabidopsis gene that is involved in the control of cell differentiation in leaves. Planta 214 694–702. [DOI] [PubMed] [Google Scholar]
- Sussex, I.M. (1954). Experiments on the cause of dorsal ventrality in leaves. Nature 174 351–352. [Google Scholar]
- Sussex, I.M. (1955). Morphogenesis in Solanum tuberosum L: Experiment investigation of leaf dorsoventrality and orientation in the juvenile shoot. Phytomorphology 5 286–300. [Google Scholar]
- Tang, G., Reinhart, B.J., Bartel, D.P., and Zamore, P.D. (2003). A biochemical framework for RNA silencing in plants. Genes Dev. 17 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij, F.E., Jones, L., and Baulcombe, D.C. (2002). Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 14 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret, H., Beclin, C., and Fagard, M. (2001). Post-transcriptional gene silencing in plants. J. Cell Sci. 114 3083–3091. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H., Vazquez, F., Crata, P., and Bartel, D.P. (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volbrecht, E., Reiser, L., and Hake, S. (2000). Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene. Development 127 3161–3172. [DOI] [PubMed] [Google Scholar]
- Volpe, T.A., Kinder, C., Hall, I.M., Teng, G., Grewal, S.I.S., and Martienssen, R.A. (2002). Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297 1833–1837. [DOI] [PubMed] [Google Scholar]
- Waites, R., and Hudson, A. (1995). phantastica: A gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121 2143–2154. [Google Scholar]
- Xu, L., Xu, Y., Dong, A., Sun, Y., Pi, L., Xu, Y., and Huang, H. (2003). Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 130 4097–4107. [DOI] [PubMed] [Google Scholar]
- Xu, Y., Sun, Y., Liang, W., and Huang, H. (2002). The Arabidopsis AS2 gene encoding a predicted leucine-zipper protein is required for the leaf polarity formation. Acta Bot. Sin. 44 1194–1202. [Google Scholar]
- Zgurski, J.M., Sharma, R., Bolokoski, D.A., and Schultz, E.A. (2005). Asymmetric auxin response precedes asymmetric growth and differentiation of asymmertic leaf1 and asymmetric leaf2 Arabidopsis leaves. Plant Cell 17 77–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z. (2004). amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels. Plant Cell Physiol. 45 369–385. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.