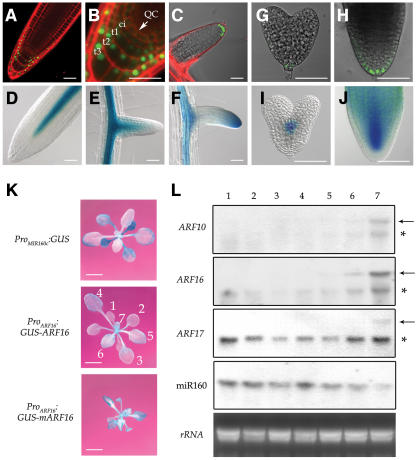
Figure 4.
Regulation of ARF16 by miR160.
(A) and (B) ProARF16:GFP-ARF16 root, showing ARF16 expression in distal stem cells and root cap cells and the localization of the GFP-ARF16 fusion protein (green) in the nucleus.
(C) The GFP signal provided by ProARF16:GFP-ARF16 was also present in distal cells of the emerging lateral root.
(D) and (E) ProMIR160c:GUS root, showing miR160 expression in the vascular bundle of primary (D) and emerging lateral (E) roots.
(F) ProARF16:GUS-mARF16 root, showing miR160-uncoupled ARF16 (mARF16) expression in both the root cap region and the vascular bundle.
(G) and (H) In developing embryos, ARF16 (ProARF16:GFP-ARF16) was expressed in the basal-most region (root cap region) at the heart (G) and bent cotyledon (H) stages.
(I) and (J) miR160 (ProMIR160c:GUS) was detected in the heart-stage embryo (I), and this miRNA then spread throughout the embryo except for the embryonic root cap region (J).
Bars = 50 μm in (A) to (J).
(K) Expression of ProMIR160c:GUS, GUS-ARF16, and GUS-mARF16 in leaves of 20-d-old plants. Numbers in the middle panel refer to the order of leaves produced from the plant. Bars = 1.0 cm.
(L) RNA gel blot analysis of ARFs and miR160 expression in leaves. Lanes 1 to 7 correspond to the order of developing leaves as shown in (K). Full-length transcripts of the ARF genes were detected only in younger leaves, whereas miR160 was highly accumulated in older leaves. Arrows and asterisks indicate the full-length and 3′ cleaved products of each mRNA.