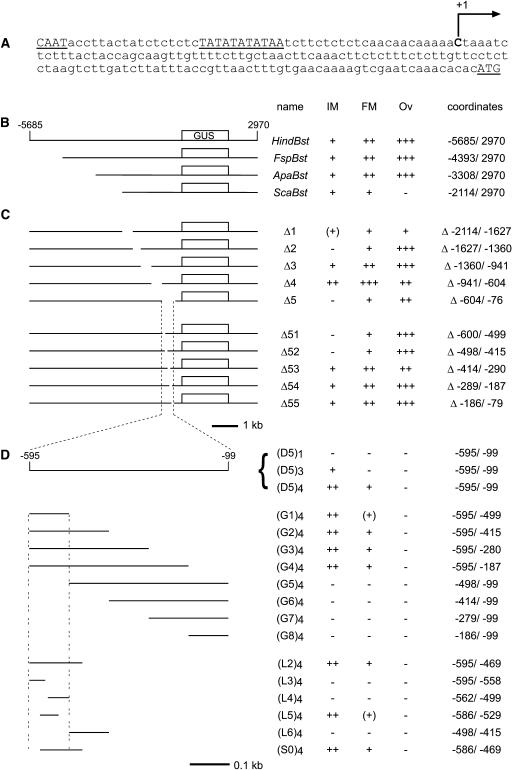
Figure 1.
Diagram of the Reporter Constructs Used in the WUS Promoter Analysis.
(A) The putative WUS transcription start site (+1) was determined by RACE PCR. Putative CAAT and TATA boxes and the start codon are underlined.
(B) to (D) For each construct, a scheme is shown at left. At right, the name, the relative staining intensities in the inflorescence meristem (IM), the floral meristem (FM), and the ovule (Ov) [−, none; (+), very faint; +, weak; ++, moderate; +++, strong], and the exact coordinates of the WUS promoter fragments or deletions (Δ) are given. The WUS coding region was replaced by the GUS coding sequence (box).
(B) Diagram of the truncation constructs analyzed.
(C) Diagram of the internal deletion constructs analyzed.
(D) Diagram of the constructs used during the functional definition of cis-regulatory sequences. To generate the reporter constructs, the monomers were multimerized as indicated and fused to -60 CaMV:GUS. D (deletion), G (gain of function), L (little deletion), and S (linker scanning) denote series of constructs.