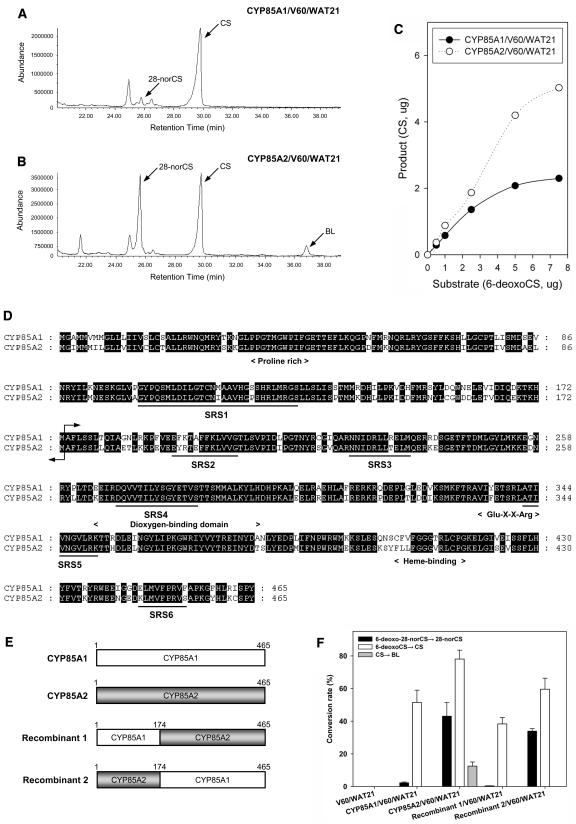
Figure 2.
Biochemical Analysis of CYP85A1 and CYP85A2 Enzyme Activity.
(A) and (B) Total ion chromatograms of the products of CYP85A1 (A) and CYP85A2 (B) heterologously expressed in the transformed yeast strains CYP85A1/V60/WAT21 and CYP85A2/V60/WAT21. The same amount of 6-deoxo-28-norCS and 6-deoxoCS was simultaneously fed to each yeast strain.
(C) Comparison of BR C-6 oxidase activity in the CYP85A1/V60/WAT21 and CYP85A2/V60/WAT21 strains. Varying concentrations (0.5, 1, 2.5, 5, and 7.5 μg) of 6-deoxoCS as substrate were fed to the two yeast strains.
(D) Alignment of amino acid sequences of CYP85A1 and CYP85A2. The conserved domains (Pro-rich, dioxygen binding, Glu-X-X-Arg motif, and Heme binding) of most Cyt P450 enzymes are indicated below the aligned sequence. Black lines indicate the six substrate recognition sites (SRSs) in Cyt P450 (Gotoh, 1992; Werk-Reichhart and Feyereisen, 2000; Williams et al., 2004). The arrow refers to the recombination site of CYP85A1 and CYP85A2.
(E) Schematic illustration of chimeric enzymes generated by exchanging the N-terminal region of CYP85A1 and CYP85A2.
(F) Change in the substrate specificity of the CYP85A1 and CYP85A2 chimeric enzymes. No metabolite was detected in the V60/WAT21 strain (control).